The fungus that made itself at home
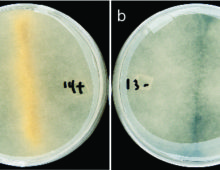
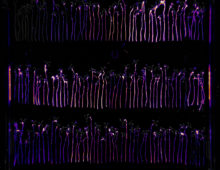
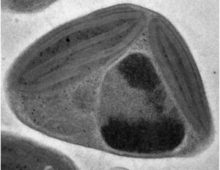
Retracing how the dry rot Serpula lacrymans adapted to a new ecological habitat. The Science By comparing genetic information from similar organisms, researchers have gained insights on why the dry rot (Serpula lacrymans) is so destructive in houses. A study involving six brown rot fungi reveals the genomic changes Serpula lacrymans has undergone in adapting… [Read More]