A fungus relies on bacteria to regulate key components of its reproductive machinery
The Science
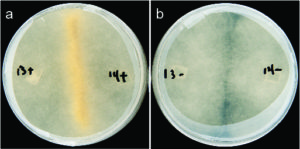
Sexual reproduction in Rhizopus microspores: (a) Successful mating between fungi harboring bacteria; (b) Lack of sex between mates cured of endobacteria. (Images by Stephen Mondo)
To better understand how beneficial organisms (symbionts) are transmitted between host generations, researchers investigated the role that bacteria living within a host (endosymbionts) have on fungal host reproduction, and the reproductive genes they regulate. The bacterial endosymbiont, Burkholderia, is recognized as a mutualist, where both species of organism benefit from association, but was predicted to have evolved from a parasitic interaction with their soil fungus host, Rhizopus microsporus. Researchers found that endobacteria establishing control over reproduction was a likely key to this evolutionary transition. Using this model, researchers also generated the first transcriptomic dataset of sexual reproduction in early fungi and discovered genes that are critical for this process.
The Impact
In the absence of the Burkholderia endobacteria, the Rhizopus fungus cannot act as a plant pathogen, it cannot reproduce asexually, and researchers have now discovered that endobacteria also regulate its sexual reproduction. The work sheds light on a poorly understood group of oleaginous or oil-producing fungi, and the impact this mutualistic interaction has on these potential large-scale biodiesel producing fungi.
Summary
In heritable mutualisms, hosts pass on beneficial symbionts between generations. The origin of this relationship though, is often antagonistic and the parasite first needs to secure its own transmission before working with the host. Using the mutualistic relationship between the plant pathogenic fungus Rhizopus microsporus (Rm) and Burkholderia endobacteria, a collaborative effort led by researchers at Cornell University and scientists at the Joint Genome Institute, a DOE Office of Science User Facility was conducted to understand how the antagonistic-to-mutualistic transition occurs. Rhizopus is a fungal pathogen of crops, including candidate bioenergy feedstocks sunflower, and maize, and a part of the oil-producing Mucoromycotina group, about which little is known. The Cornell team cultivated and experimented with the fungi and bacteria, while the DOE JGI team sequenced and annotated a host genome (Rm ATCC 52813) as part of the 1000 Fungal Genomes project.
As reported in the November 29, 2017 issue of Nature Communications, the team found that the fungus is highly dependent on the Burkholderia endobacteria to proliferate both sexually and asexually. This dependence is consistent with the addiction model of mutualism evolution; in this case, the endobacteria control the expression of ras2-1, a gene crucial to reproductive development, making the fungus reliant on the continued presence of the bacteria. By studying the Rm-Burkholderia symbiosis model, the team was able to reconstruct the reproductive pathways in several branches of the fungal kingdom, generating the first transcriptomic dataset of sexual reproduction in early fungi to find sex relevant genes across fungi. They also uncovered candidate genes, conserved across all Mucoromycotina, that appear to be involved in identifying pheromones that are critical for this fungal reproductive pathway.
Without the mutualistic Rm-Burkholderia relationship, Rhizopus is neither a plant pathogen nor able to reproduce with ease. So far, all endosymbionts discovered in fungi have been shown to substantially impact host lipid metabolism. As these oil-producing fungi are potential sustainable sources of alternative fuels, so understanding fungal-bacterial relationships can shed light on how these interactions influence lipid production in Mucoromycotina and their potential for industrial use.
BER Contact
Daniel Drell, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
daniel.drell@science.doe.gov
PI Contacts
Igor Grigoriev
Fungal Program Head
DOE Joint Genome Institute
ivgrigoriev@lbl.gov
Teresa Pawlowska
Plant Pathology & Plant-Microbe Biology
Cornell University
tep8@cornell.edu
Funding
This research was supported by the National Science Foundation and the National Institutes of Health. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Publication
- Mondo SJ et al. Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat Commun. 2017 November 29. doi: 10.1038/s41467-017-02052-8.
Related Links
- Nature Microbiology Behind the Paper: “Bacteria help fungi have sex”
- 1000 Fungal Genomes Initiative: https://jgi.doe.gov/our-science/science-programs/fungal-genomics/1000-fungal-genomes/
- JGI Fungi Data Portal MycoCosm: https://genome.jgi.doe.gov/programs/fungi/
- JGI Science Highlight: Of Mutualism and Lipid Metabolism in Fungi https://jgi.doe.gov/mutualism-lipid-metabolism-fungi/
- JGI News Release: Finding A New Major Gene Expression Regulator in Fungi https://jgi.doe.gov/finding-major-gene-expression-regulator-fungi/
- Stephen Mondo at the 2017 Genomics of Energy & Environment Meeting: http://bit.ly/JGI2017Mondo
- JGI News Release: Fungal Enzymes Team Up to More Efficiently Break Down Cellulose https://jgi.doe.gov/fungal-enzymes-team-up-efficiently-break-down-cellulose/
- JGI Publication: Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. http://dx.doi.org/10.1111/1462-2920.13669