Researchers report prevalent DNA base modification in the earliest fungal lineages
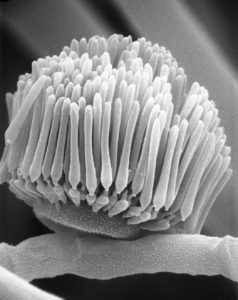
The genome of the Linderina pennispora, a fungus belonging to the earliest branches of the fungal kingdom, was sequenced and annotated as part of the Nature Genetics study. (ZyGoLife Research Consortium, Flickr, CC BY-SA 2.0)
Just four letters – A, C, T, and G – make up an organism’s genetic code. Changing a single letter, or base, can lead to changes in protein structures and functions, impacting an organism’s traits. In addition, though, subtler changes can and do happen, involving modifications of the DNA bases themselves. The best-known example of this kind of change is a methylation of the base cytosine at the 5th position on its carbon ring (5mC). In eukaryotes, a less-well known modification involves adding a methyl group to base 6 of adenine (6mA).
In the May 8, 2017 issue of Nature Genetics, a team led by scientists at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, report the prevalence of 6mA modifications in the earliest branches of the fungal kingdom. Though fungi have been around for a billion years and collectively are capable of degrading nearly all naturally-occurring polymers and even some human-made ones, most of the species that have been studied belong to just two phyla, the Ascomycota and Basidiomycota. The remaining 6 groups of fungi are classified as “early diverging lineages,” the earliest branches in fungal genealogy. They comprise a little-explored realm of fungi, providing a repertoire of important and valuable gene products for DOE missions in bioenergy and environment.
The Importance of Exploring Understudied Lineages

Watch Stephen Mondo talk about the identification and proposed role of 6mA in early diverging fungi at the 2017 Genomics of Energy & Environment Meeting: http://bit.ly/JGI2017Mondo.
“By and large, early-diverging fungi are very poorly understood compared to other lineages. However, many of these fungi turn out to be important in a variety of ways,” said study first author and DOE JGI analyst Stephen Mondo. “Consider the Neocallimastigomycetes – these fungi are one of the most powerful degraders of plant biomass currently known and have a tremendous arsenal of plant cell wall degrading enzymes which may be useful for bioenergy production. They are a good example of how exploring these understudied lineages leads to valuable biological and technological insights.”
Many of the fungal genomes used in the study were sequenced as part of the DOE JGI’s 1000 Fungal Genomes initiative aimed at producing at least one reference genome for every family of fungi. For the study, the team used 16 fungal genomes sequenced at the DOE JGI using the Pacific Biosciences sequencing platform. While the technology was used with the goal of attaining very high quality genome assemblies, DOE JGI scientists have now additionally taken advantage of this sequencing platform to explore epigenetic (5mC, 6mA) modifications. They discovered very high levels of 6mA in fungi, where up to 2.8% of all adenines were methylated, confirming these findings using multiple independent methods. The previous record holder for genomic 6mA, noted Mondo, is the alga Chlamydomonas reinhardtii (sequenced and annotated by the DOE JGI), in which just 0.4% of adenines were methylated.
6mA a Marker of Expression
“This is one of the first direct comparisons of 6mA and 5mC in eukaryotes, and the first 6mA study across the fungal kingdom,” said DOE JGI Fungal Genomics head and senior author Igor Grigoriev. “6mA has been shown to have different functions depending on the organism. For example, in animals it is involved in suppressing transposon activity, while in algae it is positively associated with gene expression. Our analysis has shown that 6mA modifications are associated with expressed genes and is preferentially deposited based on gene function and conservation, revealing 6mA as a marker of expression for important functionally-relevant genes.”
In addition to 6mA performing what seems to be the opposite role of 5mC (which suppresses expression), the team found that the presence of 5mC and 6mA are inversely correlated. Specifically, while 5mC is found at repetitive regions of the genome, the methylated adenines were clustered into dense “methylated adenine clusters” (MACs) at gene promoters. 6mA was also found consistently on both strands of DNA, which may enable propagation of methylation through cell division.
“Using genomics, we explore the diversity of fungi to develop catalogs of genes, enzymes, and pathways – parts lists for bio-based economy and bioenergy applications,” said Grigoriev. “A lot of this is encoded in early diverging fungi. In these fungi, we found that majority of expressed genes have 6mA MACs. Thus, the discovery of DNA methylation in early diverging fungi helps the research community better understand regulation of genes that encode the parts for bio-based economy and bioenergy applications.”
Mondo described the identification and proposed role of 6mA in early diverging fungi at the 2017 Genomics of Energy & Environment Meeting. Watch his talk on the DOE JGI’s YouTube channel at http://bit.ly/JGI2017Mondo.