2022 marks the JGI’s 25th anniversary. Over the next few months, we’ll be revisiting a number of notable achievements that showcase our collaborations and capabilities to enable great science that will help solve energy and environmental challenges.
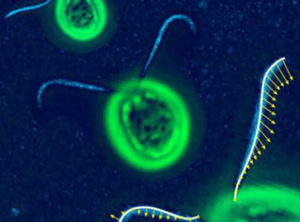
Chlamydomonas is an insightful little alga that holds clues to leveraging other organisms for sustainable biofuel production. (Image: MPI-CBG, Dresden)
Chlamydomonas is a single-celled alga with a unique place in evolutionary history. Retaining ancestral features of both plant and animal cells, it shares nearly 7,000 genes with other organisms (a third of those shared by both humans and flowering plants). Its genome also contains hundreds of genes uniquely associated with carbon dioxide capture and the generation of biomass, making Chlamydomonas (affectionately known as “Chlamy” to those who study it) a powerful reference organism for studying photosynthesis and cell motility. It’s also a model for studying lipid production in other algae that may be more viable for sustainable biofuel production.
The JGI published a partial sequence of Chlamydomonas reinhardtii in 2003; then in 2007 the full genome was sequenced and published in the journal Science. At the time, the project was co-led by Daniel Rokhsar, head of the JGI’s Computational Biology Program, along with JGI Computational Scientist Simon Prochnik, Arthur Grossman of the Carnegie Institution, and Sabeeha Merchant of UCLA. Merchant is now at UC Berkeley and also a faculty scientist in the Berkeley Lab Biosciences Area.
Merchant’s fascination with Chlamy began in her early days as a Harvard postdoc. While she had intended to work on cyanobacteria, fate stepped in when a fire at the lab relegated the experimental scientist to library research for three months. While reading up on photosynthesis, she stumbled upon an interesting observation in Chlamy. Most plants require the nutrient copper for photosynthesis; Chlamy does not. When the lab reopened, Merchant would go on to work on research that identified signaling pathways in Chlamydomonas.
So, Merchant was primed to participate when the JGI launched its effort to produce a full sequence of the Chlamydomonas genome in the early-2000s. She and other researchers came together for what was then called a JGI Jamboree, a sort of genomic hackathon to annotate a genome.
(Hear more from on Chlamy from Sabeeha Merchant in this episode of JGIotas, a Genome Insider podcast minisode.)
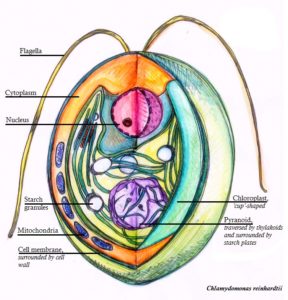
(Image credit: Ninghui Shi via Wikimedia Commons)
Since the JGI’s early sequencing of C. reinhardtii became available, data mined by the JGI shows that those sequences have been cited in almost one-fourth (23.8%) of publications focused on that specific algae. The reference genome is cited in roughly 10% of all since-released publications on green algae.
“The Chlamy genome launched over a dozen of new Chlamydomonas genome-related Community Science Program projects of all kinds,” noted Igor Grigoriev, head of the JGI’s Fungal & Algal Program, including work with Olivier Vallon of the French National Centre for Scientific Research on the Chlamy pan-genome and Marina Cvetkovska of the University of Ottawa on the multi-omics of arctic-growing Chlamy.
Merchant continues to work on the tiny alga with a mighty impact. Her lab has sequenced four additional green algal genomes using Chlamy as a reference for functional annotation, including Chromochloris zofingiensis, which has applications in both biofuel and pharmaceuticals.
The team at Merchant’s lab also discovered the existence of polycistronic genes within Chlamy, meaning that its RNA can produce up to four proteins, which has far-reaching implications.
“This is important because the textbook dogma is that in eukaryotes, one protein is produced from one RNA,” Merchant said.
Rory Craig, a former JGI intern now a postdoc in Merchant’s lab who is first author on an upcoming paper, discovered that in the original strain, a protein called RECQ3 helicase is entirely deleted, potentially resulting in many large mutations. And this is interesting because in humans (where it’s called RECQL5), the loss of this protein is associated with cancers.
“So, it shows that Chlamydomonas has this related protein that’s also causing DNA damage,” Merchant said, “that something is wrong with how DNA is being repaired in the organism.”
The upcoming paper also features a streamlined inventory of Chlamydomonas gene names. Sean Gallaher, also with Merchant’s lab and second author on the paper, studied viral genetics and gene therapy before self-training himself as a computational biologist. He spearheaded the manual curation of the Chlamydomonas genes.
The need for streamlining those gene names speaks to the impact of this tiny, single-celled organism as a model system.
“People name genes based on similarity to the Chlamy genes,” Merchant explained. “So we wanted to make sure that the Chlamy genes all had a good, proper name and that it was going to stand the test of time.”
Related links:
- JGIota: The Little Alga That Could
- Chlamydomonas reinhardtii genome on Phytozome and PhycoCosm
- JGI Blog Post: Green Algae Reveal One mRNA Encodes Many Proteins
- JGI News Release: Green Alga Genome Project Catalogs Carbon Capture Machinery and Reveals Identity as Ancient Cousin of Land Plants and Animals
- The original sequence: Science