JGI’s 25th Anniversary
The year 2022 marks the JGI’s 25th anniversary. We revisited a number of notable achievements that showcase our collaborations and capabilities to enable great science that will help solve energy and environmental challenges.
The JGI mural references dozens of iconic JGI contributions to advancing the frontiers of environmental and energy genomics. Commissioned for the JGI’s 25th anniversary and dedicated at the 2022 Annual Meeting, the two-story mural at the Integrative Genomics Building was painted by Berkeley-based artist Nigel Sussman.
 |
Behind the JGI Mural
Berkeley-based muralist Nigel Sussman on creating the JGI’s 25th anniversary mural. |
 |
Timelapse: Painting the Mural
Footage of Nigel Sussman painting the JGI mural at the Integrative Genomics Building. |
Notable stories from the past 25 years
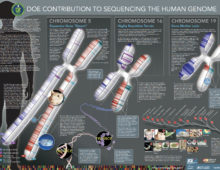 |
The Human Genome Project, or the JGI’s Origin Story
In 1997, the U.S. Department of Energy (DOE) united the people and resources from three national labs to work on the Human Genome Project. The JGI contributed ~12% of the total human genome. |
 |
Historical perspectives on JGI’s 25 years At the 2022 JGI Annual Meeting, speakers shed light on how the JGI was established, all that it has contributed, and what they’re excited about in the JGI’s future. |
 |
Poplar, The Bioenergy Tree
In 2006, JGI collaborated on the first tree genome, Populus trichocarpa. The genome offers exciting opportunities beyond just a bioenergy feedstock crop. |
 |
Pioneering Metagenomics Research Paves Path for Nobel
A conversation on extremophiles elements with a longtime JGI collaborator piqued the interest of Jennifer Doudna, leading to her 2020 Nobel Prize for co-developing CRISPR-Cas9 technology. |
 |
Roots of a Mutualist Relationship
The genome sequence of Laccaria bicolor, a forest fungus in symbiosis with poplar, was published in 2008. Today ~200 mycorrhizal fungal genomes are publicly available on the fungal portal MycoCosm. |
 |
A Single Cell, Myriad Microbial Discoveries Single cell genomics offers a way for researchers to study the planet’s microbial diversity, much of which remains unknown and uncultured. |
 |
Studying Sorghum’s Survival Skills
Since publishing the sorghum reference genome, the JGI continues collaborating to better understand its toughness and energy storage. |
 |
Solving the Mystery of the Missing Oil
Working with Berkeley Lab researchers, the JGI’s single cell genomics and metagenomics capabilities were harnessed for a systems biology approach to the Deepwater Horizon oil spill. |
![Chlamydomonas is an insightful little alga that holds clues to leveraging other organisms for sustainable biofuel production. [Image: MPI-CBG, Dresden]](https://jgi.doe.gov/wp-content/uploads/2022/08/Chlamy.LeadImage-300x222.jpg) |
The reference Chlamydomonas genome is cited in roughly 10% of all since-released publications on green algae. |
 |
Building a Better Bean
The common bean genome has fueled further research into disease tolerance in crops, stressors and nitrogen fixation. |
 |
Fueling Investigation into Methane-Making Microbes
Supporting studies involving methane-making microbes in ruminants to drive solutions for stemming methane-emissions. |
 |
Strengthening Soybean
Aside from supporting work on the soybean plant, the JGI has also helped researchers understand a fungus threatening these crops.
|
 |
Former JGI interns share how these experiences have influenced their career journeys. |
 |
Cow Rumen and the Early Days of Metagenomics A 2011 Science paper has led to a hands-on undergraduate research course at four universities. |
 |
Following Fungi that Pry Apart Plant Polymers
Gut fungi from livestock may be able to provide tools beyond biomass breakdown, extending into biochemical production. |
 |
Mapping Switchgrass Traits with Common Gardens
Long-term research investments in switchgrass genomics are helping the grasses become a key player in biomass-based fuels. |
 |
Team Science to Build Communities around Data
Researchers are forming communities to generate high quality genomes with the JGI. |
 |
Expanding Metagenomics to Capture Viral Diversity
The JGI has leveraged their work as metagenomics pioneers to boost understanding of environmental viruses. |
 |
Tracking & Subduing the Plague of California’s Oak Woodlands
Overseeing engagement activities to control the spread of sudden oak death. |
