Uncovering associations between plant roots and fungi that can help or harm plant host health.
The Science
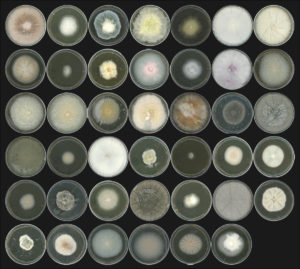
Photograph of 41 fungal isolates representative of the A. thaliana root mycobiome (© S. Hacquard / MPIPZ)
Researchers compared the genomes of fungi that colonize Arabidopsis thaliana roots with genomes from other plant-associated fungi. Their analysis identified fungal gene families that help determine if these fungi act as friends or foes.
The Impact
Microbial communities comprised of fungi and bacteria can thrive in plant roots. They play key roles in maintaining the plant’s health and diversity, which can in turn boost yields for both food and for candidate bioenergy feedstock crops.
Summary
A plant is like an office building, its roots are akin to the ground floor lobby. People walking into the lobby have many reasons for being there, but they’re not always immediately apparent.
Similarly, the microbes found in plant roots may have unknown roles. Some plants and fungi have mutually beneficial (e.g., mycorrhizal) relationships, but not all plants and fungi can form such symbiotic partnerships.
To learn more about these other plant-fungal relationships, an international team led by graduate student Fantin Mesny in Stéphane Hacquard’s group at the Max Planck Institute for Plant Breeding Research (MPIPZ) studied how fungi associate with roots in A. thaliana plants. The work also involved researchers at the French National Research Institute for Agriculture, Food and Environment (INRAE) and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab). Their findings were described in a recent Nature Communications paper.
The work was partly enabled by a JGI Community Science Program (CSP) proposal led by INRAE’s Francis Martin that contributed toward filling gaps on the fungal Tree of Life through the 1000 Fungal Genomes Project. Hacquard’s group had assembled a mycobiome culture collection consisting of fungal species from natural Arabidopsis populations around Europe. The JGI sequenced and annotated 41 fungal isolates from the collection. The team then compared this dataset against nearly 80 previously published fungal genomes on the JGI data portal MycoCosm. They looked for genes known to be involved in fungal-host interactions and identified 84 gene families that were common in fungi that live within plants (endophytes). These include genes that degrade plant cell walls and break down plant sugars.
The results also revealed that the most beneficial fungi in root mycobiomes weren’t necessarily the most abundant members of these communities. In contrast, many of the fungi that dominate the mycobiome were predicted to evolve from ancestors that were likely harmful to plants and can efficiently colonize roots by degrading the plant cell wall component pectin. The results therefore predicted that these root-colonizing fungi must be kept in check by the host and other microbes not to impede host health.
For JGI Fungal Program head Igor Grigoriev, the paper is an “excellent example” of how 1000 Fungal Genomes project can enable studies of plant root microbiome and other microbiomes by providing reference genomes of the key community participants and enabling future metagenomics studies.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
PI Contact
Stéphane Hacquard
Max Planck Institute for Plant Breeding Research
hacquard@mpipz.mpg.de
Funding:
The sequencing project was funded by the U.S. Department of Energy (DOE) Joint Genome Institute, a DOE Office of Science User Facility, and supported by the Office of Science of the U.S. DOE under Contract No. DE-AC02-05CH11231 within the framework of CSP 1974 “1KFG: Deep-sequencing of ecologically relevant Dikaria”. This work was supported by funds to S.Hac from a European Research Council starting grant (MICRORULES 758003), the ‘Priority Programme: Deconstruction and Reconstruction of the Plant Microbiota (SPP DECRyPT 2125)’ and the Cluster of Excellence on Plant Sciences (CEPLAS), both funded by the Deutsche Forschungsgemeinschaft. F.M. salary was covered by the DECRyPT 2125 programme. This research was also supported by the Laboratory of Excellence ARBRE (ANR- 11-LABX-0002-01), the Region Lorraine, the European Regional Development Fund, and the Plant–Microbe Interfaces Scientific Focus Area in the Genomic Science Program, the Office of Biological and Environmental Research in the US DOE Office of Science (to F.M.M.). M.K. acknowledges funding from the SLU Centre for Biological Control (CBC). The Austrian Science Fund FWF project P30460-B32 is acknowledged for funding L.A.
Publication:
- Mesny F et al. Genetic determinants of endophytism in the Arabidopsis root mycobiome. Nat Commun. 2021 Dec 10;12(1):7227. doi: 10.1038/s41467-021-27479-y.
Related Links:
- Nature Communications Behind the Paper blog: Fungi in plant roots: friends, enemies or frenemies?
- MPIPZ Release: Differentiating friends from foes in the fungal root microbiome
- JGI Community Science Program
- JGI 1000 Fungal Genomes Project
- JGI CSP Proposal: Deep Sequencing of Dikarya
- JGI fungal portalMycoCosm