Nanohaloarchaeota cultures reveal they are symbionts and not free-living organisms.
The Science
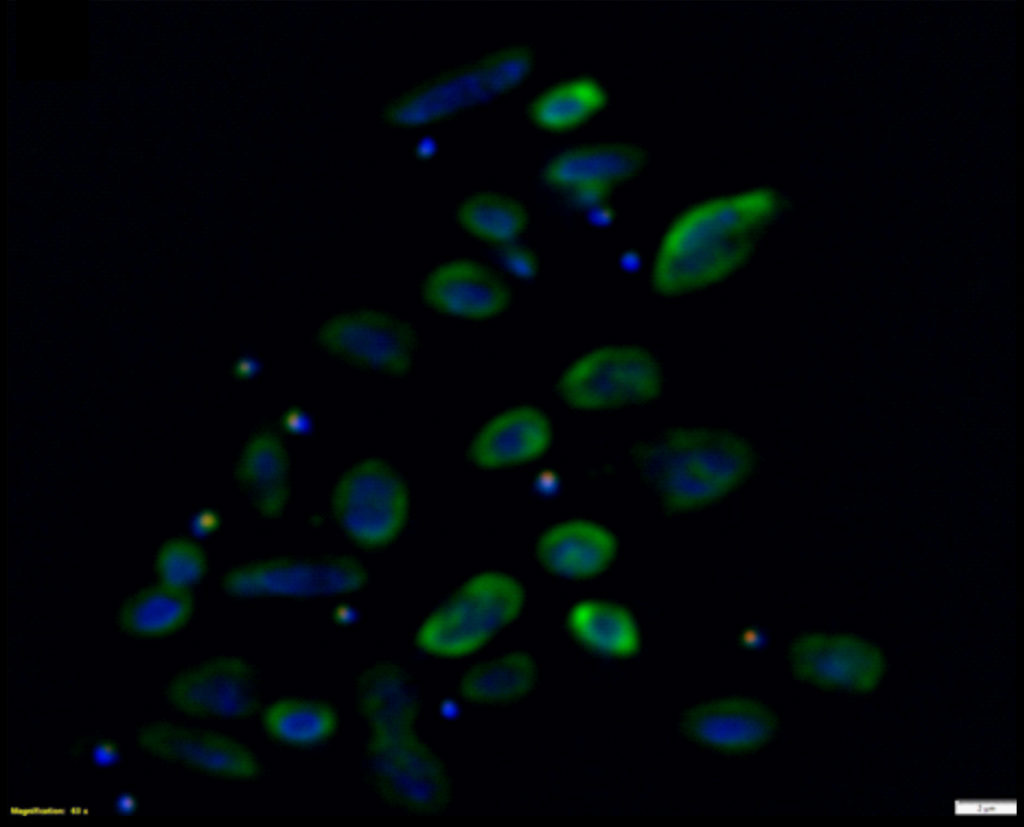
FISH of Nha-C enrichment with Hrr. lacusprofundi ACAM34-hmgA. Fluorescence micrograph shows individual Nha-C cells amongst Hrr. lacusprofundi cells. Nha-C cells labelled with a Cy5 (red fluorescence) conjugated probe; Hrr. lacusprofundi cells labelled with a Cy3 (yellow fluorescence, recolored to green to improve contrast) probe; all nucleic-acid containing cells stained with DAPI (blue fluorescence). Composite image of all three filters. Scale bars represent 2 µm. (Josh Hamm, UNSW)
Researchers employed multiple microbiology and ‘omics techniques to experimentally determine that Nanohaloarchaeota are symbionts, rather than free-living organisms as had been originally thought.
The Impact
The Antarctic lakes are a “treasure trove” of unknown microbes that play critical roles in environmental processes (related to interactions of soil-sediment environments, water and air-atmosphere with the humans and the biosphere), including the global carbon cycle. The JGI is sequencing large numbers of microbes and microbial communities that contribute to carbon cycling, and is interested in understanding the genomic basis and ecological consequences of microbe-microbe interactions. Using metagenomes recovered directly from environmental samples and cultivated microbes, researchers have been able to gain further insights about how microbes have adapted to thrive in polar ecosystems. The work by Cavicchioli’s team sheds light on the range of archaeal symbiotic lifestyles found in the Antarctic lakes and offers a genetically-tractable model system that could help researchers better understand the factors controlling microbial symbiotic relationships.
Summary
Symbiotic relationships are long-term biological interactions between one or more biological organisms. One hallmark of an obligate symbiont—entirely dependent on the other for required nutrients—is genome size. The obligate symbiont’s genome is often reduced compared to that of a free-living organism.
First discovered in 2011, Nanohaloarchaea are part of the DPANN (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaea) superphylum of extremophilic archaea first outlined by JGI scientists and their collaborators in 2013. While most DPANN lineages are predicted symbionts, the Nanohaloarchaeota were predicted to be capable of autonomous growth because they had larger genome sizes and their cells were not observed to associate with host cells.
In the Proceedings of the National Academy of Sciences, a team led by Rick Cavicchioli at the University of New South Wales along with researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility, experimentally demonstrated that Nanohaloarchaeota are not free-living archaea but rather symbionts.
Cavicchioli’s graduate student, Joshua Hamm, successfully cultured two strains of Nanohaloarchaeota and showed they required cell-cell contact with the Antarctic haloarchaeon Halorubrum lacusprofundi. To do this, water samples were collected from the Rauer 1 and Club lakes in East Antarctica and enriched for native microbes. Metagenomes from enrichment cultures were sequenced and assembled by the JGI. From these assemblies, complete or near-complete population genomes were recovered—termed “metagenome-assembled genomes” (MAGs)—of the nanohaloarchaea strains Nha-R1 and Nha-C, which Hamm cultured.
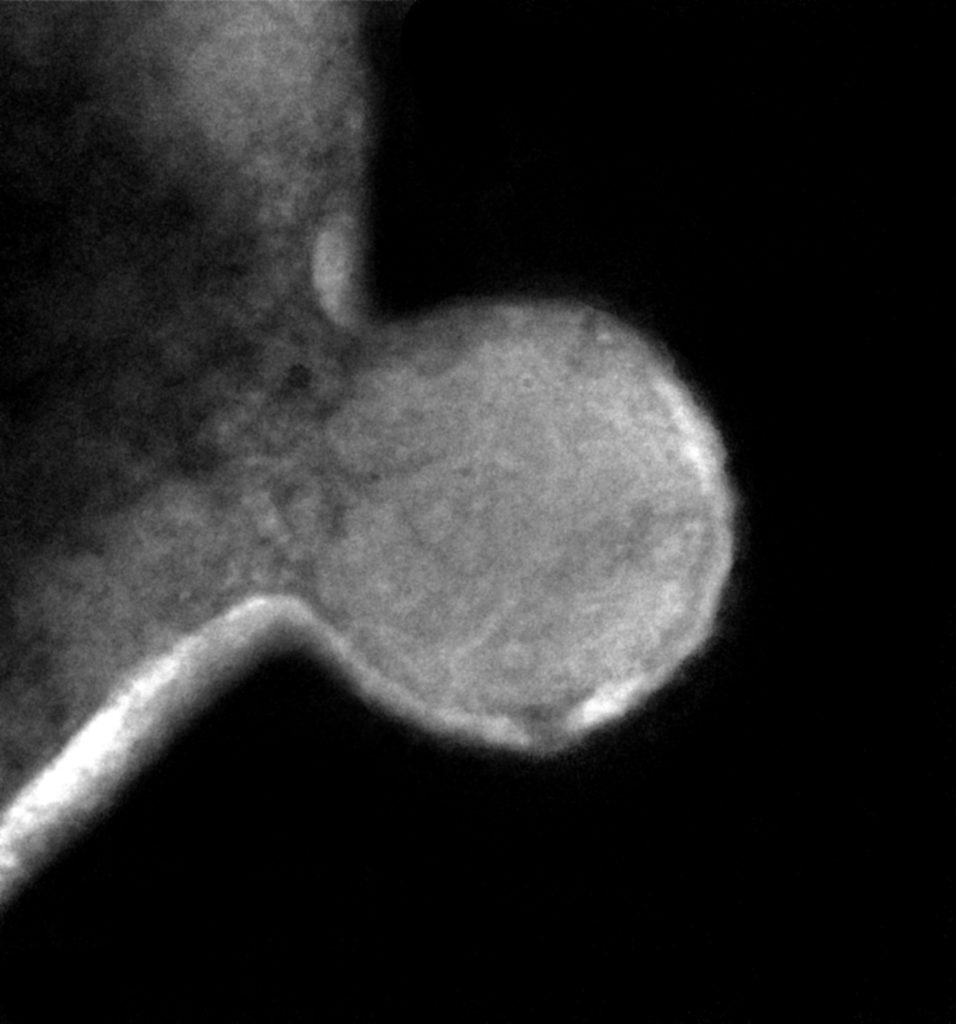
TEM of Nha-C FACS cells with Hrr. lacusprofundi ACAM34. A single ‘budded’ cell (putative Nha-C) intimately associated with a larger cell (putative Hrr. lacusprofundi). (Josh Hamm, UNSW)
The researchers then pursued the Nha-C strain for further study, tagging the cells with individual fluorescent probes. Co-fluorescence suggested the Nha-C cells were interacting with another abundant species in the enrichment cultures, Hrr. lacusprofundi cells, the first member of the Archaea isolated from Deep Lake, the coldest place on Earth known to support life, and previously sequenced by the JGI. Transmission electron microscopy (TEM) provided high resolution images confirming the Hrr. lacusprofundi were directly transferring cytoplasmic content to Nanohaloarchaea.
Using flow cytometry, the team purified Nha-C cells from the enrichment cultures and showed that the Nanohaloarchaeota grew only when cultivated with Hrr. lacusprofundi. The team also found that the Nanohaloarchaeota cannot produce certain essential parts of a cell: amino acids, co-factors, and membrane lipids, suggesting they rely on Hrr. lacusprofundii for them. One of the most distinctive features of the Nanohaloarchaeota was discovered with proteomic analyses. The species harbors genes encoding SPEARE proteins—which contain serine protease, adhesion and restriction endonuclease domains—of up to 8,500 amino acids in length. These were actively synthesized intact in the Nha-C cells. The most conserved region in the SPEARE proteins appears to be involved in DNA degradation, and these proteins may be directly involved in enabling the Nanohaloarchaeota to gain access to Hrr. lacusprofundi host cells to obtain essential nutrients to maintain the symbiotic relationship.
The combination of multiple microscopy and omics techniques enabled the team to closely study the nature of the Nanohaloarchaeota species’ symbiotic relationship.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
PI Contact
Rick Cavicchioli
School of Biotechnology and Biomolecular Sciences
UNSW Sydney
r.cavicchioli@unsw.edu.au
Funding:
This work was supported by the Australian Research Council (DP150100244) and the Australian Antarctic Science program (project 4031). The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under contract no. DE-AC02-05CH11231. S.E. was supported by the EMBO Long-Term Fellowship ALTF 188 –2014, which is co-funded by the European Commission (EMBOCOFUND2012, GA-2012-600394) and supported by Marie Curie Actions.
Publication:
- Hamm JN et al. Unexpected host-dependency of Antarctic Nanohaloarchaeota. Proc Natl Acad Sci U S A. 2019 Jun 28. pii: 201905179. doi: 10.1073/pnas.1905179116
Related Links:
- Nature Research Microbiology Community “Behind the Paper”: Discovery, hypothesis & Koch’s postulates – Nanohaloarchaeota are NOT free-living
- JGI Community Science Program
- JGI News Release: Boldly Illuminating Biology’s “Dark Matter”
- CSP 2015 Proposal: Seasonal variation in Antarctic microbial communities: ecology, stability and susceptibility to ecosystem change
- JGI Science Highlight: Defining a Pan-Genome for Antarctic Archaea
by Massie S. Ballon