DOE JGI researchers sequenced and analyzed a cyanobacterium with known nitrogen-fixing capabilities.
The Science:
Researchers sequenced and analyzed the uncommonly large genome of a nitrogen-fixing cyanobacterial strain, comparing it to the more than 150 extant cyanobacterial genomes.
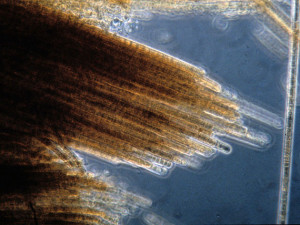
The golden-brown surface mats that indicate a Trichodesmium erythaeum bloom have led to them being called “sea sawdust.” (Image by FWC Fish and Wildlife Research Institute via Flickr CC BY-NC-ND 2.0)
The Impact:
Unlike many of the existing cyanobacterial genome sequences, the genome of Trichodesmium erythaeum IMS 101 is large compared the number of genes encoded. Understanding the significance of conserving the non-coding regions could help researchers understand how these algae with their unusual genomic architectures, can thrive in nutrient-poor regions.
Summary
In the global oceans, cyanobacteria such as Prochloroccus and Synechococcus are critical because despite their small size, their sheer abundance involves them in the fixation of nearly half of total atmospheric carbon captured annually and its conversion into organic material through photosynthesis. For this reason, several projects conducted at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science user facility managed by Lawrence Berkeley National Laboratory, focus on understanding the mechanisms involved in photosynthesis, as well insights into the roles they play in other global cycles. One such project, conducted under the DOE JGI’s Community Science Program, was called CyanoGEBA and focused on sequencing 50 phylogenetically diverse cyanobacteria genomes.
Published ahead online March 23, 2015 in the Proceedings of the National Academy of Sciences, a consortium including DOE JGI researchers sequenced and analyzed the genome of a strain of the cyanobacteria Trichodesmium erythraeum isolated off the coast of North Carolina. Unlike Synechococcus and Prochlorococcus, this cyanobacterium plays a key role in fixing atmospheric nitrogen. The absence of this nutrient can be a limiting growth factor.
DOE JGI researchers have previously collaborated on studies that indicated that some cyanobacteria selectively reduce their genomes to thrive in nutrient-poor environments. In the case of the Trichodesmium genome however, the team found that just under two-thirds of the total DNA was predicted to encode proteins and of the remaining non-coding DNA, nearly 90 percent was expressed as RNAs. Additionally, this non-coding DNA contained “numerous” DNA repeat sequences. The team compared the genome of this T. erythaeum strain to a draft genome of another T. erythaeum strain isolated from the Tropical Atlantic, and a partial draft of a different Trichodesmium species isolated in Hawaii. They found that all three Trichodesmium strains had similarly low gene densities along with large genomes. One suggestion offered by the team was that the noncoding regions might contain regulatory elements required for the proper expression of needed genes for growth, both in the lab and in the wild.
Another theory they proposed was, “that the intergenic regions experience gradual inflation during certain evolutionary intervals characterized by [algal] bloom-driven selective sweeps.” Unlike other cyanobacteria, Trichodesmium can thrive both in large clusters, or with other microbes. In the latter situation, their shared co-existence could lead to multiple Trichodesmium subpopulations that could contribute to the non-streamlined genomes seen.
The architecture of Trichodesmium genomes is unexpected and consistent across widely diverse habitats; genomic architecture of other cyanobacteria is very different, with higher gene densities and less non-coding DNA. The Trichodesmia fix nitrogen while most other cyanobacteria do not. The relationship of Trichodesmium’s “extra” DNA and this important capability, central to understanding microbial ecology, remains to be worked out.
Contact
Tanja Woyke
DOE Joint Genome Institute
twoyke@lbl.gov
Funding
- U.S. Department of Energy Office of Science
- National Science Foundation
- University of Southern California
Publication
- Walworth N et al. Trichodesmium genome maintains abundant, widespread noncoding DNA in situ, despite oligotrophic lifestyle. Proc Natl Acad Sci U S A. 2015 Mar 23. pii: 201422332. [Epub ahead of print] doi: 10.1073/pnas.1422332112
Related Links
- http://newscenter.lbl.gov/2003/08/14/genomes-of-tiny-microbes-yield-clues-to-global-climate-change/
- http://news.usc.edu/80277/dna-of-bacteria-crucial-to-the-ecosystem-defies-explanation/http://jgi.doe.gov/why-sequence-cyanobacteria-synechocystis-transcriptome/
- http://jgi.doe.gov/carboxysomes-and-the-carbon-cycle/
- http://jgi.doe.gov/doe-jgi-science-highlights-cyanobacterial-genomics-a-basis-for-improvements-in-photosynthesis/
- http://jgi.doe.gov/doe-jgi-science-highlight-benefits-of-diversity-driven-genome-sequencing-in-cyanogeba/
- http://jgi.doe.gov/single-species-reveals-myriad-genomic-variations/
- http://jgi.doe.gov/tracing-ancestral-responses-light-algae-plants/
- http://jgi.doe.gov/streamlining-common-survival-strategy-marine-microbes/
- http://jgi.doe.gov/the-inner-workings-of-a-bacterial-black-box-caught-on-time-lapse-video/