Visualizing a novel, candidate viral genome found in the deep subsurface ecosystem.
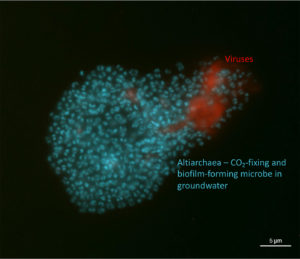
Image of biofilm with both Altiarchaea (blue) and viruses (red). (Victoria Turzynksi and Lea Griesdorn)
The Science
Altiarchaea are carbon-fixing microbes and targets of multiple viruses in Earth’s deep subsurface. They are abundant representatives of deep subsurface ecosystems. A team of researchers described how the viruses repeatedly attempted to infect and destroy the host archaea – and how the microbes resist. The battle waged below the Earth’s surface is reconstructed by combining a study of microbial communities (metagenomics) and a chemical labelling approach.
The Impact
Altiarchaea are abundant and dominant populations in oxygen-poor environments without oxygen deep below ground. These archaea fix carbon and nitrogen, but the viruses that infect Altiarchaea cause the cells to burst. This action triggers subsurface carbon cycling, releasing fresh organic carbon which can serve as a food source for other microbes into the environment.
Summary
In J. R. R. Tolkien’s “The Lord of the Rings,” the Hornburg fortress had withstood all attacks – until the Battle of Helm’s Deep. The army of the dark wizard Saruman repeatedly tried to break through, finally resorting to explosives to succeed.
Altiarchaea, like the Hornburg, have a history of countering multiple attempts at virus infections. Their genomes include short DNA sequences that code for CRISPR systems, which help bacteria resist foreign genetic elements by incorporating short sequences from infecting viruses and phages. In these sequences are virus fragments called spacers, that indicate repelled attempts to infect and lyse the microbial cells.
This demonstration of the virus-host arms race in uncultivated yet abundant subsurface organisms was recently reported in Nature Communications. The work was co-led by Victoria Turzynski in Alexander Probst’s lab at the University of Duisburg-Essen and included researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab).
To learn more about how microbial populations respond to virus infections, researchers are using Altiarchaea as a model system to study virus-host interactions. Approximately 150 archaeal viruses had been previously identified. The JGI researchers surveyed publicly available data in the JGI’s IMG/M database, information the Probst lab used to search for and find 13 additional predicted viral genomes. The team dubbed these viruses Altivir, finding them in three of the four subsurface ecosystems known to host Altiarchaea.
The team then tracked how Altiarchaea build immunity to viruses. To do so, they compared samples collected from an aquifer in 2012 and 2018 against a biofilm sample collected in 2018. They looked for and found CRISPR spacer sequences in the samples that corresponded to eight separate groups of viruses.
To visualize one particular viral genome in action, they utilized a targeted technique called virusFISH. Using fluorescently tagged probes, the team was able to visualize the behavior of a particular candidate virus under a microscope. The viruses were adsorbed by the cells, which then burst, spreading the virions out and moving the carbon within the microbial community. These findings also suggest that lytic viruses are more common in deep biosphere microbial communities than had been thought. Lysogenic viruses, which prefer to infect the host cell and then get passively replicated with the host DNA, have long been considered to be more abundant in these environments.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Alexander Probst
University of Duisburg-Essen
[email protected]
Funding:
This work received funding by the Alfred P. Sloan foundation (grant number G-2017-9955), the Ministry of Culture and Science of North Rhine-Westphalia (Nachwuchs-gruppe “Dr. Alexander Probst”), and the NOVAC project of the German Science Foundation (grant number DFG PR1603/2-1). The authors acknowledge sampling logistics provided by the University of Regensburg, i.e., by Harald Huber and Sebastien Ferreira-Cerca, and sequencing of MSI metagenomes within the Census of Deep Life Sequencing call 2018, phase 14 project “Development of novel archaeal viruses and the corresponding CRISPR arrays of a highly abundant carbon fixer in Earth’s crust”. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231.
Publication:
- Rahlff J and Turzynski V et al. Lytic archaeal viruses infect abundant primary producers in Earth’s crust. Nature Communications. 2021 July 30. doi:10.1038/s41467-021-24803-4
Related Links:
- University of Duisburg-Essen Release: A Food Chain in the Dark
- JGI Release: Tracking Microbial Diversity Through the Terrestrial Subsurface
- JGI Release: Boldly Illuminating Biology’s “Dark Matter“
- JGI Highlight: Metagenomics Leads to New CRISPR-Cas Systems
- JGI Highlight: Mining Metagenomes for Cas Proteins