Got 12 Gb of free storage to download it all?
To better understand how Earth’s vast and diverse microbial population helps regulate global nutrient cycles, it helps to understand how viruses infect microbes, and affect their functions and metabolic processes. Two years ago, even though the number of viruses is estimated to be at least two orders of magnitude more than the microbial cells on the planet, sequence databases held less than 2,200 sequenced DNA virus genomes, compared to the approximately 50,000 bacterial genomes on file. That ratio changed dramatically when JGI researchers unveiled more than 125,000 partial and complete viral genomes, described in an August 2016 Nature paper, and boosted the number of known viral genes 16-fold.
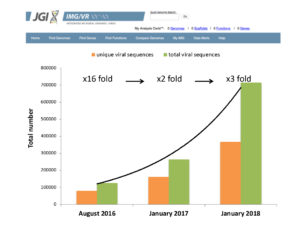
Growth rate of virus identification and microbial host prediction for the new release of IMG/VR database. Growth over time in the total and unique number of viral sequences in IMG/VR. (Image composite by David Paez-Espino).
On the heels of that release, the JGI launched IMG/VR, describing it then in the January 2017 “Database” issue of Nucleic Acids Research as the largest publicly available database with isolate reference DNA viruses and computationally identified contiguous viral sequences (“contigs”) from thousands of ecologically diverse metagenomics samples. By then, the contents of the IMG/VR database had doubled from the figure referenced in August 2016.
This month, IMG/VR unveiled its largest release of viral datasets yet, with a year’s worth of data mined from metagenome sequences both by the JGI and externally submitted, and isolates sourced from the National Center for Biotechnology Information (NCBI). There are now over 715,000 viral datasets, with nearly 8,500 isolate viruses, and over 700,000 viral contigs.
“The viral diversity in IMG/VR has tripled since January 2017,” said JGI bioinformaticist David Paez-Espino. “We have now identified over 34,000 viral sequences targeting several microbial taxa for the very first time, we’ve associated new viruses to known microbial genomes, and the vast majority of the gene content (over 15 million genes in total) remains hypothetical or unknown, meaning that there is tremendous potential for new discoveries out of that gene pool.”
“The idea is to release all the viral data we could find along with key information, such as samples in which a virus was identified, and what its expected host might be, so that the community can efficiently analyze and mine these data through this integrated viral genomics resource,” said JGI scientist Simon Roux.
As detailed in an August 2017 Nature Protocols report, the data generation was made possible by a semi-automated discovery pipeline. Paez-Espino adds that in addition to the data release, IMG/VR has a new feature that allows researchers to download the entire database. “You can search IMG/VR for homologous viruses if you provide your sequences, or search for a host using our spacer BLAST database, but now you can also bulk download the whole database with metadata associated.” Despite the massive data release, he notes, downloading more than 715,000 viral contigs and the rest of the database would take up just 12 GB of storage on a computer. That’s up from the database size of 4GB had it been downloadable a year ago.
“We anticipate that the release of such massive viral sequence data to the research community will drive novel discoveries and understanding of the viral world,” said Prokaryote Super Program head Nikos Kyrpides, “We invite the community to explore these data and provide feedback on how we can improve IMG/VR.”
Hear how researchers are already mining the IMG/VR database at the upcoming Viral EcoGenomics & Applications (VEGA) Symposium to be held March 14-15, 2018 during the JGI’s 13th Annual Genomics of Energy and Environment Meeting at the Hilton San Francisco Union Square. Additionally, on March 13, the Prokaryote Super Program hosts a workshop detailing the microbial and metagenomics resources and capabilities available to users through partnering with the JGI – and how current collaborators are developing technologies to further innovate – and harnessing the Integrated Microbial Genomes suite of tools. Learn more at usermeeting.jgi.doe.gov/vega.
Through the Community Science Program (CSP), the JGI has launched a New Investigator call for proposals, with the emphasis on providing pilot data to form the foundation of a large-scale CSP proposal submission. Proposals are due by March 1, 2018, and must be independent of ongoing accepted proposals. Additionally, the lead PIs on these New Investigator proposals cannot have been lead PI on any previously accepted JGI CSP or FICUS proposal. The New Investigator calls for proposal will go out four times a year, and replace the CSP Small Scale Calls – click here to learn more.
References:
- IMG/VR portal: https://img.jgi.doe.gov/vr
- Bulk Download the IMG/VR database: https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html
- JGI News Release: Unveiled: Earth’s Viral Diversity
- JGI Science Highlight: DOE JGI Database of DNA viruses and retroviruses debuts on IMG platform
- JGI Publication: “Uncovering Earth’s virome” http://rdcu.be/FDVo
- JGI Publication: “IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses” https://doi.org/10.1093/nar/gkw1030
- JGI Publication: “Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data” https://www.nature.com/articles/nprot.2017.063