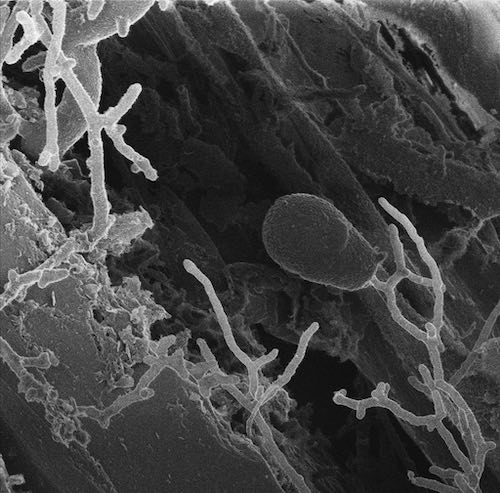
The anaerobic fungus Anaeroromyces robustus growing on reed canary grass. (Vaithiyalingam Shutthanandan, PNNL/EMSL)
Fungi from herbivore poop make surprisingly complex compounds — an energetically expensive feat thought to be too costly for them.
The Science
Anaerobic fungi, which die in the presence of oxygen, dwell in herbivore guts and help them digest their last leafy meal. In their evolutionary history, these fungi branched off early from aerobic fungi, which can breathe oxygen — just like we do. Oxygen is a rich source of energy, and because anaerobic fungi can’t harness it, scientists long held that these fungi don’t have the energy to make complex compounds called natural products. Yet, combing through the genomes of four anaerobic fungal species has revealed, for the first time, that this group is unexpectedly powerful: they can whip up dozens of complex natural products, including new ones.
The Impact
In the past, natural products have been an important source of antibiotics. Indeed, these anaerobic fungi may be using these compounds for self-defense. But the compounds may also be involved in how the fungi metabolize plant biomass, which could inform how scientists can more easily convert plants into carbon-neutral biofuels. What’s more, many natural products have potential to be used as precursors for polymers and commodity chemicals, as well as drop-in biofuels. This work shows that anaerobic fungi might be a new source of novel compounds to help bolster the bioeconomy.
Summary
A team led by Michelle O’Malley, a chemical engineer at the University of California (UC), Santa Barbara, collected the droppings of a sheep and a goat at the Santa Barbara Zoo, and those of a horse in Massachusetts, in their hunt for anaerobic fungi. By growing extracts without oxygen in ruminant fluid — which smells terrible —they were able to isolate four strains: Anaeromyces robustus, Caecomyces churrovis, Neocallimastix californiae, and Piromyces finnis. In work reported in 2017, the fungal genomes were sequenced by the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory. More recently, O’Malley’s team has begun to understand the outsized role the fungi play in digesting plant material in herbivore guts.
A deeper dive into the genomes and follow-up experiments, recently reported in the Proceedings of the National Academy of Sciences, has now revealed that the fungi also produce a number of natural products.
In the lab, O’Malley and her team — notably, first author Candice Swift — verified that the fungi expressed biosynthetic gene clusters, which are “building blocks” of natural products. The team then confirmed that these expressed genes translated into expected proteins. That work was done in collaboration with the Environmental Molecular Sciences Laboratory (EMSL), a DOE Office of Science user facility at Pacific Northwest National Laboratory. The team’s research at both the JGI and EMSL was enabled by the Facilities Integrating Collaborations for User Science (FICUS) initiative, which allows researchers to more easily integrate the expertise and capabilities of more than one DOE Office of Science National User Facilities, with the stroke of a single proposal.
The next step for the team was to try to identify what natural products the fungi were making. So, Trent Northen, head of the JGI Metabolomics Technology Group, with scientists Ben Bowen and Katherine Louie conducted mass spectrometry experiments (or analyses) to break up the compounds and to puzzle out their structures. Their results indicate that anaerobic fungi synthesize molecules that are sources of new chemical diversity; the team identified what looked to be three groups of novel compounds, as well as (potentially) one previously known natural product: baumin. Baumin is an antioxidant, previously known to be made only by aerobic fungi.
In order to identify more of the fungal natural products, O’Malley and her team plan to use genetic tools to boost production and isolate the compounds. Although what compounds they make is still largely a mystery, anaerobic fungi clearly have unexpected chemical potential.
Contacts:
BER Contacts
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
Paul Bayer
Program Manager, EMSL
Climate and Environmental Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Paul.Bayer@science.doe.gov
PI Contact
Michelle O’Malley, Ph.D.
Professor, Department of Chemical Engineering
University of California, Santa Barbara
momalley@ucsb.edu
Funding:
This research project was supported by a grant from Santa Barbara Cottage Hospital and the University of California, Santa Barbara and a gift from the Gordon and Betty Moore Foundation to MAO. A portion of this research was performed under the JGI-EMSL Collaborative Science Initiative and used resources at the DOE Joint Genome Institute and the Environmental Molecular Sciences Laboratory, which are DOE Office of Science User Facilities. Both facilities are sponsored by the Office of Biological and Environmental Research and operated under Contract Nos. DE-AC02-05CH11231 (JGI) and DE-AC05-76RL01830 (EMSL). CLS is also supported by a National Science Foundation Graduate Research Fellowship Program under Grant No. 1650114, a UCSB Graduate Division Dissertation Fellowship, and the UCSB Connie Frank Fellowship. The authors acknowledge support from the Center for Scientific Computing from the CNSI, MRL: an NSF MRSEC (DMR-1121053) and NSF CNS-0960316.
Publications:
- Swift CL et al. Anaerobic gut fungi are an untapped reservoir of natural products. PNAS. doi: 10.1073/pnas.2019855118
Related Links:
- Engagement Webinar on FICUS: How to Apply for Resources at Multiple DOE User Facilities
- LBL Science Snapshot: The Green Secrets of Goat Poop
- News Release: Fungal Enzymes Team Up to More Efficiently Break Down Cellulose
- News Release: Biofuel Tech Straight From the Farm
- JGI’s Natural Prodcast podcast: Primer on Natural Products
- JGI’s Fungal & Algal Program
Byline: Alison F. Takemura