What if we understood plants and how they adapt to their ever-changing environments better? We could unlock new innovations to drive more productive food, medicine, and bioenergy crops. But most available genomes are from narrow swaths of the plant tree of life. One project aims to change that. Relevant links are linked throughout and at the end.
The Joint Genome Institute presents the Genome Insider podcast.
Transcript of the episode
ALISON: Hey! I’m Alison Takemura, and this is Genome Insider, a podcast of the US Department of Energy Joint Genome Institute, or JGI. Plants permeate our lives. Maybe you’ve worn a cotton shirt recently. Or plopped on a couch with a wooden frame under all of those cushions. Or maybe you just ate a meal — a meal that I bet would not have been possible without plants. Pretty much all animals depend on plants whether directly or indirectly

Hong Ma, plant biologist at Penn State University, is working to understand the evolution of land plants. The gourds beside him are an example of their richness. (Alison F. Takemura)
HONG: You know, think of a tiger – it’s going to eat maybe a wild deer but the deer graze on grass.
ALISON: That’s Hong Ma, a plant biologist at Penn State. And like other animals, we humans can’t get far without plants. Here’s Hong’s colleague at Penn State, plant biologist Claude dePamphilis.
CLAUDE: Plants are fundamental to humans. People use plants for all kinds of things. We use plants, for food, for shelter for materials. Basically, everything we do depends on plants. Try to go through a day without something made from a plant and you’ll find you just can’t.
ALISON: Given the significant role plants play in our lives and in earth’s ecosystems, scientists really want to get into the weeds of how plants do all the things that they do. That’s why Hong and Claude are part of a team that’s undertaken a big project to deepen our understanding of plants— especially branches on the plant tree of life that have been mostly ignored. The project is called Open Green Genomes; Green genomes because they come from plants, and Open because the project adheres to a model of open science. Why is this undertaking important?
DOUG: We really want to understand what plants are all about; all processes from photosynthesis to making diverse chemicals, medicines, crops.
ALISON: That’s Doug Soltis, a plant biologist at the Florida Museum of Natural History at the University of Florida. He’s one half of a plant research power couple, the other member being plant biologist Pam Soltis at the Florida Museum of Natural History. With plants, there’s a lot to explore. Scientists estimate that there are more than 400,000 land plant species on earth. Here’s Pam.

Pam Soltis and Doug Soltis, plant biologists at the University of Florida, are also part of the Open Green Genomes project. (Alison F. Takemura)
PAM: What is it that makes all of that diversity? How do they adapt to different environments? How do they produce the structures that they produce?
ALISON: For example, how do they make wood?
PAM: How do they produce the chemicals that they produce? All of that is encoded in their genomes, and by understanding a bit more about their genomes, we can start to make progress toward understanding what’s responsible for all of the diversity that we see around us.
ALISON: Pam, Doug, Hong, and Claude are among 120 scientists in the Open Green Genomes project. The project aims to sequence the genomes of 35 new plants. These plants have been carefully selected to better inform our understanding of what makes plants tick and how their adaptations have evolved.
What’s the current status with plant genomes? Well, right now, most of the hundreds of available plant genome sequences are of plants with economic importance, like food and potential bioenergy crops, or they’re plant model systems, like Arabidopsis thaliana, which is a petite flowering plant with small leaves that, to some, look like mouse ears. But, as Doug points out,
DOUG: That still leaves a huge chunk of the green plant tree of life to cover. That’s our goal in this project: by looking at mosses and ferns and various green algal lineages and also conifers to get a really broad picture of green plant evolution.
ALISON: How different, exactly, are plants? If you imagine a redwood and aloe vera, well, you start to get a picture, right? But here’s another measure: If you were to randomly pick any two plants being sequenced in the Open Green Genomes project, they’d probably be more different than any two mammals. The whole of mammalian diversity — dwarfed by that of plants. Plant genomes can also be radically different from one plant to another. Take for instance, size. Here’s Claude again.

Claude dePamphilis, plant biologist at Penn State University, is one of more than 100 principal investigators working on the Open Green Genomes project to fill in the plant tree of life. (Alison F. Takemura)
CLAUDE: Plants vary from having a genome that’s just a few percent of the size of the human genome to others that are as much as 50 or more times the size of a human genome. Every biochemical process that we can think of varies significantly in terms of the genes that encode them in plant genomes, and every one of these important agricultural or biofuels-related or wood-development related processes can’t really be understood through the study of just one or a few carefully studied models.
ALISON: Gathering more information from more genomes really helps — for one, because we don’t know what a lot of genes do. We just don’t have that kind of functional understanding yet. Here’s Hong. Oh! And, note, there were some children playing at the arboretum where I did this recording, so you’ll hear them in the background.
HONG: The difficulty with any organism is that the functional studies are very slow, very time consuming, very difficult sometimes. And there’s a lot of people who try hard to understand even a single organism. But we still, for example, Arabidopsis is probably the most intensively studied at the functional level. But I think we don’t even know the function of 10 percent of the genes. So the other 90 percent, we know the sequence, we know a lot of their expression patterns, but we don’t have definitive evidence they’re important for a particular process.
ALISON: But with more genomes comes more opportunities to compare and try to figure out what genes underpin these economically important processes. For example, how plants make wood. Comparing genes is essentially comparing parts lists. So you can ask, what genes do all woody plants have that non-woody plants don’t. Maybe some of those genes are responsible for forming wood.
Here’s a different example of how studying the genetic richness in plants has already been helpful. It comes from studying the cacao tree, Theobroma cacao. These trees produce cacao fruit, which we ferment into cocoa, the star ingredient of chocolate. Claude looks across cacao and its relatives to analyze disease resistance in cacao.
CLAUDE: The relatives of cacao give us a lot of insight in how the disease resistance genes have evolved. And gives many clues that we’ve actually been able to put into practice in identifying real genes that really affect disease resistance and susceptibility in this crop plant of cocoa.
ALISON: The Open Green Genomes project could produce more insights like Claude’s work in cacao — just on a much broader scale. And the Joint Genome Institute is providing support. Here’s Claude.
CLAUDE: Joint Genome is uniquely positioned to be able to do a project like this, because they have the technology, and the knowledge, and bioinformatic skill to handle this diverse collection of genomes.

Jim Leebens-Mack leads the ambitious Open Green Genomes project to sequence 35 new plant genomes. (Courtesy of the University of Georgia)
ALISON: The project was started in 2018 by Jim Leebens-Mack, a plant biologist at the University of Georgia. Of the 35 target plant genomes, Jim says…
JIM: We’ve got five of those that are 100 percent complete. And data has been put in the databases and scientists around the world are accessing those data and using them to address a very wide range of questions about, about the biology of their plants of interest.
ALISON: One of the primary data repositories for these genomes is Phytozome, JGI’s plant genome database.
JIM: In writing the proposal, we pointed out that JGI’s commitment to open science and in their infrastructure for promoting open science our objective of generating genome sequences and making them available to the world as quickly as possible.
ALISON: The team is making these genomes available to everyone, as soon as the genomes pass muster, and potentially, even before getting out a manuscript. I don’t know about you, but as a trained scientist, I find that pretty unusual.
But first the scientists have to get the DNA sequenced. Getting the DNA itself, though, isn’t easy. Here’s Pam.
PAM: So we tend to think that sequencing the genome and assembling a genome is the really tricky part. But sometimes it’s actually cultivating the species so that you can get enough tissue so that you can get a good DNA extraction. And sometimes plants have so many nasty compounds in them that it’s really difficult to get a good extraction. By good extraction, we mean we want a lot of DNA. We want the DNA to be in really big pieces. We don’t want it to be sort of broken up by either the mechanics of extracting the DNA or by the chemicals that are released in the cells. So sometimes just generating that DNA, it can be really, really challenging. And one of the things that I think our whole community has found is that we have to do a lot of experimentation when you work with a new species. Maybe you have a really good approach, and you use that a lot of times and it works great then, you try it on a new species, and it doesn’t work at all. So then you’re back to the drawing board, and you have to think about what is it about that species? What is its chemistry like, where does it grow? What are its pests? Maybe all of that sort of information can help you develop some new tricks to extracting the DNA.
ALISON: That DNA then gets sent to JGI.
PAM: JGI is doing all of the sequencing and the genome analysis. And they’re really good about also helping with training, so that we can all learn more about how best to sequence a bigger complex genome, how to put the genome pieces together in a way that makes sense, and then how to interpret the results.
ALISON: That’s happening for every one of the 35 plants undertaken in this project. And while getting 35 new genome sequences is a huge accomplishment, the Open Green Genomes project is even more ambitious. To really understand how plants do what they do – photosynthesize, grow wood, fend off disease, and so on – the team is also tracking gene expression. Jim says that that means collecting a lot more samples.
JIM: We want to make sure that we’re sampling lots of different developmental stages, reproductive organs, roots, below ground organs, above ground vegetative organs, so that we can look at the expression of genes and each of these different organs or developmental stages, and use that gene expression data to help us start gaining some idea of what are the 30 to 30 to 50,000 genes that were identified in each of these genomes. What are those genes doing?
ALISON: This means that the team needs to follow the plant from seedling to maturity.
JIM: We can’t just take up a potted plant or some leaf material and generate a genome sequence and leave it at that. The other thing that would be terrible is if we got started on a particular genotype, and started sequencing its genome, and then the plant died.
ALISON: That would be tragic! To avoid an untimely demise, Jim and the team think very carefully about which plants they choose. Not only for sampling now, but also in the future.
JIM: We want to make sure that these plants are in secure living collections so that other people can go back to these particular reference plants for the genomes that we’ve sequenced, perhaps get cuttings from those plants and do additional work on those.
ALISON: Jim says that getting the genomes and gene expression data for these plants is like having more complete blueprints to work with.
JIM: By filling in that picture, it gives us access to this, this toolkit, that plants have drawn on, throughout their evolution, and do some engineering. So, you know, intentional tinkering of the genes, gene content, gene function.
ALISON: One example that you could imagine is uncovering more of the genes and gene regulators that give rise to wood production.
JIM: And, you know, using that information to build in a deliberate way, a poplar tree that is growing faster, producing better wood from a biofuels perspective, and then becoming a better biofuels crop.
ALISON: For more on using wood from poplar trees as a biofuel feedstock, check out our story “THE Bioenergy Tree” in Season 2 Episode 7.

Ceratopteris richardii, or C-fern, the first plant genome sequence of the Open Green Genomes project to be written up in a manuscript. (David Randall and Zhonghua Chen)
With the Open Green Genomes project, we’ve had a chance so far to speculate on its future fruits. But what’s happening right now? The project is actually heading toward a major milestone: scientists are preparing the project’s first manuscript, to accompany a finished genome sequence.
JIM: The genome was assembled and annotated, just beautifully by, by our collaborators at the Joint Genome Institute.
ALISON: And that genome is of a fern, called Ceratopteris, which starts with a ‘C’, richardii. It’s called C-Fern for short and is actually used as a teaching tool. (Transcript note: For more, see “Ferns: the final frond-tier in plant model systems.”)
JIM: Many folks may have played with C-Fern. I might have been exposed to it in high school biology lab, when we looked at fern reproduction.
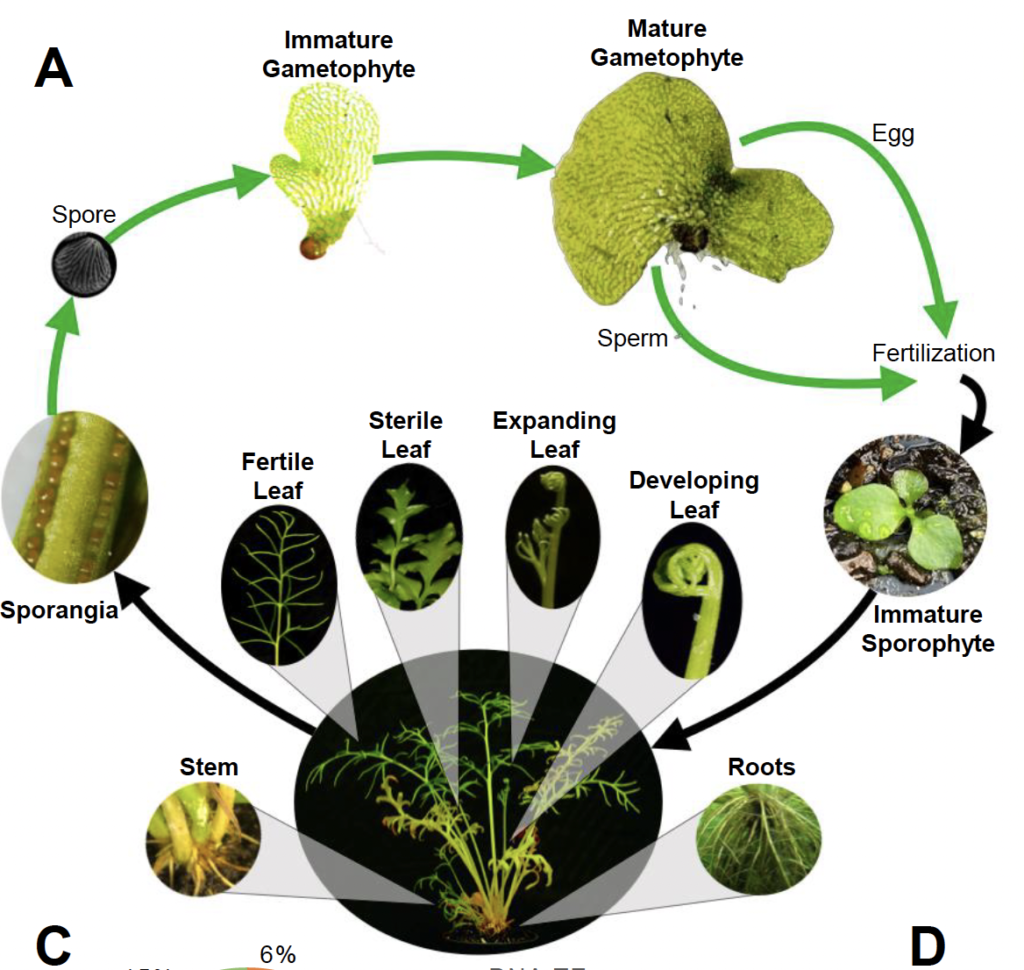
The life cycle of C-fern, a study subject for many a high-school student. (Courtesy of Jim Leebens-Mack)
ALISON: Hm, I don’t remember having that experience, unfortunately. But getting back to the genome, Jim and his team have found some interesting, unexpected features. One is that it’s big — it’s 50 times larger than the genome of the model plant Arabidopsis. Two, the genome has some gnarly content.
JIM: It’s got lots of instances where genes have been sucked up from bacteria, or fungi, incorporated in the genome, and then those genes that were first transferred by a horizontal gene transfer event, have actually duplicated, in some instances multiple times in the fern genome lineage.
These are bacterial or fungal genes that are now doing some important function in a plant.
ALISON: Weird – and cool. A third notable discovery is that, some 50-60 million years ago, the genome underwent a duplication event.
JIM: But the order in which those genes are occurring in the genome has been mixed up much more than we’ve seen in other plant genomes.
For whatever reason, in this lineage leading to C-Fern, we’ve got a very rapid rate of structural change.
ALISON: These are just the first insights expected to come from the genomes, or genome assemblies, generated in this project, which is supported by JGI’s Community Science Program. Jim had some really nice things to say.
JIM: It couldn’t be done without JGI. JGI has just been rock solid. And since the first JGI genome, the poplar genome sequence, JGI has, maintained and expanded their expertise in generating sequence data, using it to make assemblies and genome annotations, interpreting those genomes. It’s just invaluable. It’s.. this project, I wouldn’t even tried to do it, frankly, if it weren’t for the capabilities of the JGI.
ALISON: Those helpful capabilities have extended even into the management of this project, which is something my mind grappled with, because, I’ll remind you, this project involves more than 120 principal investigators.
JIM: It’s logistically a complicated project, and it requires some pretty adept project management. And is another way in which JGI’s experience and expertise has been mission critical.
ALISON: One of the key people Jim has worked with is JGI Project Manager Kerrie Barry. Wielding spreadsheets, Kerrie and other JGI staff have kept careful track of the progress for each of the plant genomes. And they’ve helped the Open Green Genomes team manage and document their monthly remote team meetings.
The JGI team of (left to right) Kerrie Barry, Dario Copetti, Jayson Talag, Jeremy Schmutz, Jane Grimwood, and Jerry Jenkins support Jim Leebens-Mack and the rest of the Open Green Genomes team. (Courtesy of Jim Leebens-Mack)
JIM: Folks at JGI, Kerrie, have been just wonderful at doing that and making it so I can go back and look at a, either a spreadsheet or our, our meeting notes from each of our meetings. And seeing okay, this is where we left off.
ALISON: Yes, indeed: nearly every successful project has strong management behind it.
Well, this is where we leave the Open Green Genomes project for now. But what can we look forward to? By setting out to sequence 35 completely new plant genomes, the Open Genomes Project is an audacious undertaking. It’s like climbing a new mountain range. But at the summits are new vistas. The project has the potential to provide unprecedented views of the genetic elements that diverse plants rely on. And these findings will help us to better understand, and even improve, the plants that we turn to for food, medicine, and bioenergy.
ALISON: This episode was directed and produced by me, Alison Takemura, with editorial and technical assistance from Massie Ballon, David Gilbert, and Ashleigh Papp.
Genome Insider is a production of the Joint Genome Institute, a user facility of the US Department of Energy Office of Science. JGI is located at Lawrence Berkeley National Lab in beautiful Berkeley, California.
A huge thanks to plant biologists Hong Ma and Claude dePamphlis at Penn State University, Doug and Pam Soltis at the Florida Museum of Natural History at the University of Florida, and Jim Leebens-Mack at the University of Georgia, for sharing their passion for plants and uncovering the secrets within plant genomes. Special thanks, as well, to a behind-the-scenes partner: the Arizona Genomics Institute, which produces the high-quality plant DNA that JGI needs for sequencing.
Because we’re a user facility, if you’re interested in partnering with us, we want to hear from you! We have projects in genome sequencing, synthesis, transcriptomics, metabolomics, and natural products in plants, fungi, algae, and microorganisms.
If you want to collaborate, let us know! Find out more at jgi.doe.gov forward slash user dash programs.
And if you’re interested in hearing about cutting edge research in secondary metabolites, also known as natural products, then check out JGI’s other podcast, Natural Prodcast.
And, dear audience, it’s with a heavy heart that I say goodbye — this is my last time hosting Genome Insider. It has been such a pleasure to bring you stories from the frontlines of environmental genomic science. I’ll miss it, and I’ll miss all of you. In this transition, Genome Insider will be taking a few months hiatus, But then it’ll re-emerge with a new host at the helm.
In the meantime, let us know what you’ve thought of Genome Insider so far! Leave us a review, tweet us @JGI, or record a voice memo and email us at JGI dash comms at L-B-L.gov. That’s jgi dash c-o-m-m-s at l-b-l dot g-o-v. We’d love to hear from you!
That’s it for now; take care!
Additional information related to the episode:
- Overview of the Open Green Genomes approved proposal
- JGI Genome Portal: plant genomes statuses of the Open Green Genomes project
- JGI plant genomics resource Phytozome
- Ceratopteris richardii genome on Phytozome
- JGI News Release: 2018 DOE JGI Community Science Program Allocations Announced, including the Open Green Genomes project
- Our contact info:
- Twitter: @JGI
- Email: jgi-comms at lbl dot gov

