Proposals encompass multiple capabilities of the national user facility
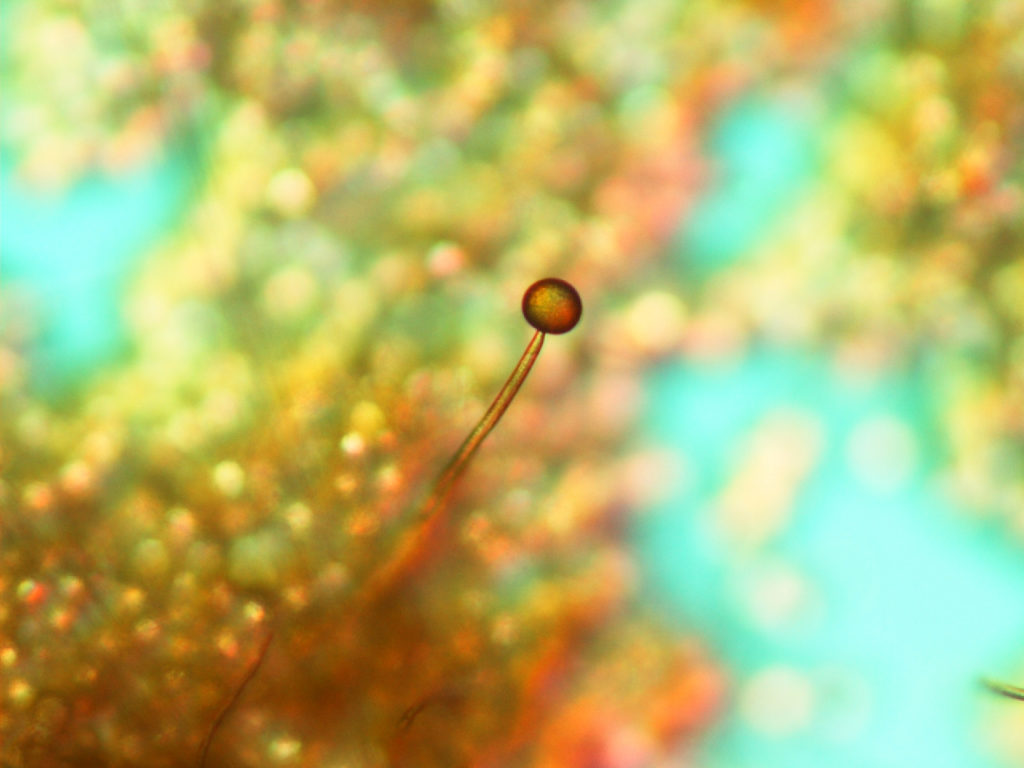
Mucor circinelloides sporangiophore. Fungal responses to DNA methylation in the context of developmental signals is just one aspect of Victoriano Garre‘s proposal. (S. Torres-Martínez. University of Murcia, Spain.)
Though organisms can be studied in isolation, a more comprehensive picture emerges when their environmental interactions are taken into account. Along the same lines, many of the 30 proposals selected for the 2018 Community Science Program (CSP) of the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, aim to utilize multiple genomic and analytical capabilities, along with scientific expertise, to users focused on the underlying mechanisms involved in bioenergy generation and biogeochemical processes.
“These new CSP projects demonstrate further growth into multi-omics, with nearly all requesting functional genomics technologies including transcriptomics, epigenomics and metabolomics, and roughly half spanning more than one of our Science Programs,” said Susannah Tringe, DOE JGI User Programs Deputy. “They bring exciting new investigators, ideas and approaches to the application of genomics to DOE mission science.”

East River in Colorado is where watershed studies are being conducted as part of a proposal from Jill Banfield. (Courtesy of Jill Banfield)
The CSP 2018 proposals were selected from 76 full submissions based on 94 letters of intent. Click here to see the full list of approved CSP 2018 proposals. Additionally, 60 percent of the accepted proposals come from new DOE JGI primary investigators.
Among this year’s accepted proposals:
- Dan Buckley of Cornell University aims to dissect soil microbial food webs using stable isotope probing, a method that traces nutrient fluxes by providing labeled substrates that are consumed and incorporated into DNA. This will allow his group to identify and characterize uncultivated microbes that have crucial roles in the soil carbon cycle.
-

Developing gene atlas maps to study grass-microbe interactions that can lead to either healthy or virus-infected Brachypodium is a partial aim of Kranthi Mandadi‘s proposal. (Courtesy of Kranthi Mandadi)
Boulos Chalhoub of the French National Institute for Agricultural Research (INRA) will explore how DNA methylation and repeated hybridization have shaped the polyploid Brachypodium hybridum, a relative of candidate bioenergy grasses such as switchgrass. Polyploid plants contain multiple sets of chromosomes, which makes sequencing and assembling these genomes more challenging, but can also make the plants more stress tolerant.
- Maureen Coleman of the University of Chicago will apply integrated ecosystem genomics across the Great Lakes, which hold 20 percent of the Earth’s surface freshwater. She will characterize the metabolic diversity and activity of microbes participating in nutrient cycling in these important aquatic systems.
-

The interactions between cyanobacteria – such as those found in Octopus hot spring biofilms at Yellowstone National Park – and viruses is the topic of Devaki Bhaya‘s proposal. (Courtesy of Devaki Bhaya)
Byron Crump of Oregon State University aims to apply metagenomics and metatranscriptomics to microbes that metabolize dissolved organic matter in river ecosystems, critical conduits for land-to-ocean transfer of materials including terrestrial organic matter.
- Alisa Huffaker of the University of California (UC), San Diego will harness the DOE JGI’s diverse capabilities in the plant, metagenome, and synthesis programs, and use metabolomics to produce systems analysis of the metabolic diversity of sorghum and maize to better understand microbiome interactions and how these grasses tolerate various stresses.
- James Leebens-Mack of the University of Georgia will develop a comparative plant genomics framework involving high-quality genome assemblies and annotations for 35 species. The Open Green Genomes Initiative, which lists 98 co-primary investigators along with Leebens-Mack, will improve comparative analyses of the genes, regulatory networks and metabolic pathways influencing plant growth and responses to environmental stress, and inform engineering of plants for efficient production of biofuels and bioproducts. In the proposal, the team noted that the large number of investigators “is a testament to the importance and broad interest in developing infrastructure – including strategically sampled reference-quality genome sequences – to aid comprehensive comparative analyses across the green tree of life.”
-

Illustration of disease symptoms caused by rust fungi, the topic of Sebastien Duplessis‘ proposal, on different host plants. (photos by M. Catherine Aime)
Udaya Kalluri of Oak Ridge National Laboratory wants to know how modifying plant cell walls in the candidate bioenergy feedstock crop poplar, one of the DOE JGI’s Plant Flagship Genomes, impacts plant-microbe interactions and the utility of poplar for biofuel production.
- Corby Kistler of the USDA-ARS Cereal Disease Lab and the University of Minnesota targets the mutually antagonistic relationships between bacteria and fungi in native prairie soil, and how their system of checks and balances allow perennials to thrive. His proposal will harness the DOE JGI’s fungal, microbial, metagenome and metabolomics capabilities.
- Yu Liu of the University of Texas Southwestern Medical Center plans to define the global regulatory network of DNA chromatin structures in the filamentous fungus Neurospora crassa, an important model organism for the fungal conversion of biomass.
- Norma Martinez-Gomez of Michigan State University aims to identify the rare earth element-dependent enzymes involved in plant-microbe interactions that can boost crop yields such as candidate bioenergy feedstocks while reducing the need for fertilizers.
-
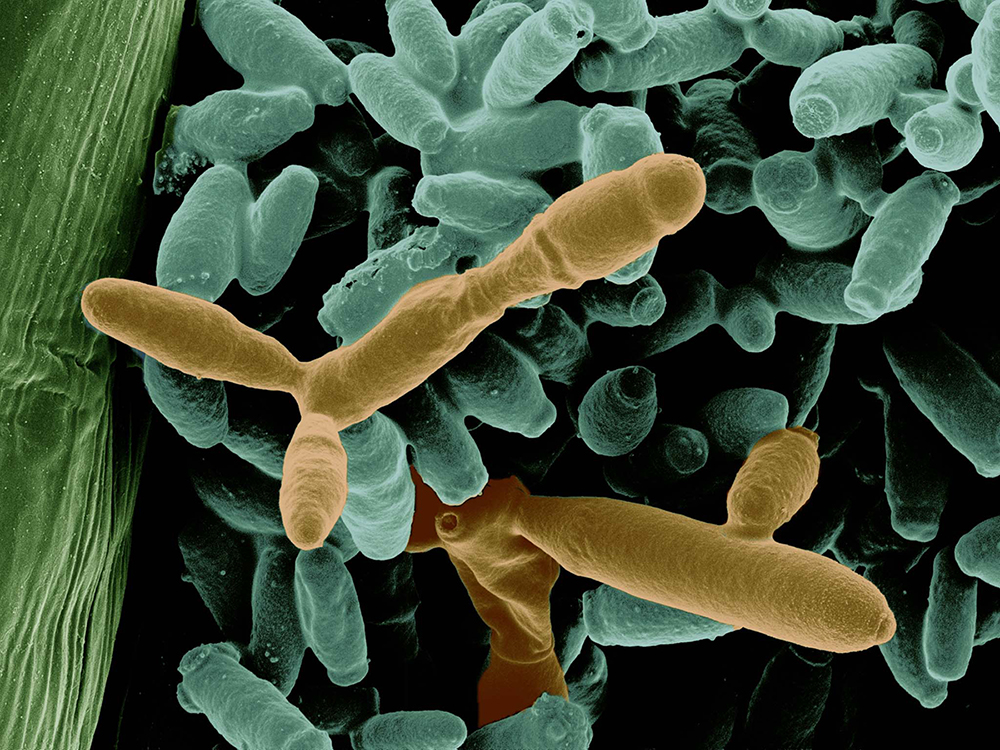
Nectar yeasts including Metschnikowia gruessii are the focus of Tadashi Fukami‘s proposal. (Manpreet Dhami, Tadashi Fukami, and Lydia-Marie Joubert)
Jennifer Martiny of UC Irvine will explore the activity of surface soil microbes on leaf litter in an arid ecosystem to understand how daily cycles in moisture and temperature impact nutrient cycling.
- Neslihan Tas of Lawrence Berkeley National Laboratory, recently named a DOE Early Career Research Program awardee, plans to use sequencing and metabolomics to explore microbial functions over time in watershed ecosystems. Her team is already collecting samples from soil-aquatic interfaces at two Colorado high-elevation streams.
- Ru Zhang of the Donald Danforth Plant Science Center is investigating how the model green alga Chlamydomonas reinhardtii – genome sequenced and assembled by the DOE JGI – regulates its responses to heat stress and day length with the aid of metabolomics. The information could lead to algal strains that produce more biomass for conversion to biofuel.

A proposal by Jana U’Ren calls for a genomic survey of Xylariaceae fungi. The morphological diversity of Xylariaceous endophytes is shown here. (J.M. U’Ren)
These projects, as well as the other 17 selected in this round of the CSP, represent new ways of incorporating DNA sequencing with other analytic approaches to explore biological relationships that underpin bioenergy and biogeochemical processes. Understanding how microbes live in association with other microbes and plants, how plant genome diversity results from DNA modifications, how microbes exploit available local nutrient resources, and how metabolic plasticity can be understood and potentially exploited, among other questions, will help scientists leverage high throughput analytic methods for DOE mission purposes.