Lawrence Livermore National Lab biologist Jennifer Pett-Ridge collaborated with JGI scientists on an ambitious project: to bring in robots to help process experiments that measure microbial activity in soil. Now, the researchers and robots have made these experiments easier for scientists everywhere.
The Joint Genome Institute presents the Genome Insider podcast.
Transcript of the episode
ALISON: Hey! I’m Alison Takemura, and this is Genome Insider, a podcast of the US Department of Energy Joint Genome Institute, or JGI.

Jennifer Pett-Ridge, senior staff scientist and leader of the Environmental Isotope Systems Group at Lawrence Livermore National Lab, in Livermore, CA. (Jacob Long)
Last time, we talked to Jennifer Pett-Ridge at Lawrence Livermore National Lab about a technique called stable isotope probing. It’s a method that uses heavier isotopes of elements, like carbon 13 instead of carbon 12, to tag active microbes. If a microbe is chowing down on something labelled with carbon 13, it’ll end up incorporating some of that carbon into its DNA. That will make its DNA much heavier. Stable isotope probing, or ‘S-I-P’, SIP, for short, is a really powerful tool for identifying microbes that are active from those that are dormant or just lounging about. The technique also just got way easier. Here’s Jennifer to explain.
JENNIFER: Over the past five years or so, I’ve just gotten more and more interested in using stable isotope probing, to be able to identify the active cells in soil and really quantify their levels of activity.
However, the, the process of doing SIP, the standard SIP, at least, where you centrifuge, and then you fractionate the nucleic acids that have been separated by, by the centrifuge. Gosh, it’s one of the most tedious laboratory procedures that, that there is.
And if you know anyone who has done it, boy. Their, their eyes will kind of glaze over when they start to tell you about it. What it used to involve is, is taking that that centrifuge tube, puncturing the bottom of it, and slowly draining out the liquid. And counting the drops, literally, you know, seven drops might equal a fraction.
And so you’re staring at this thing, counting seven drops, moving in another tube, counting another seven drops moving in another two, and then you’ve taken one sample and created maybe 70 different fractions…. it’s, it’s painful, it takes a very special person to, to do. I was lucky enough to have three of these very special people in my lab, at the same time when the call came out for the Emerging Technologies Opportunity Program.
ALISON: I’ll get back to that program in a second. But first those three very special people in Jennifer’s lab were: Ashley Campbell, Erin Nuccio, and Steve Blazewicz. Together with Jennifer, they came up with a proposal for that program that Jennifer just mentioned: JGI’s Emerging Technologies Opportunity Program. The program is a way for scientists to collaborate with JGI on technologies that could benefit not only those scientists with their proposal, but also other researchers that might work with JGI in the future: Future users. Jennifer and her team’s idea was right on the money.
JENNIFER: We pitched an idea of, well, what if we could get a robot to do several of the the, really laborious kind of steps that are painful for humans, and but, you know, are really just, it’s just repetitive, like many molecular biology tools.
ALISON: Uh, yes! Sign me up for making science less tedious.
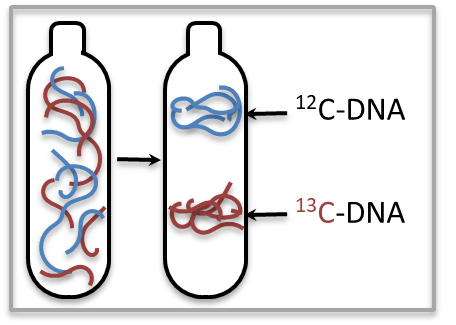
After carbon 13 is introduced to the soil, the DNA of active organisms becomes more dense, and thus heavier. Here, a tube contains mixed heavy and light DNA is centrifuged in order to separate the layers of carbon 12 and carbon 13-enriched DNA. Then, the samples are further split. Pett-Ridge’s team submitted a proposal to the JGI to make this process more automated. (Courtesy of Jennifer Pett-Ridge)
JENNIFER: And we then spent two, two and a half years developing a whole pipeline, which involves actually two different robotic steps, one that does, takes care of the dripping and counting of fractions, and another liquid handling robot that takes care of cleanup, and precipitations and desalting of samples. And we worked very closely with the JGI. Because we knew, you know, that they had really actually more expertise than we did in particularly the second kind of robot, the Hamilton liquid handling robot, they, they had worked with those for many years. But we created a whole process and just developed the metrics of how successful each step was.
ALISON: In the end, the new process slashes the amount of labor hours to about a twelfth of what they were.
JENNIFER: And the old method, we, you know, we still have colleagues who use it. And they’re running more like six or seven samples a week, and it’s exhausting. It’s not something you want to do week after week after week. Whereas with the pipeline we’ve created, we find we’re able to pretty routinely run 16 samples a week, you know, month after month. It’s a real improvement over the old approach, and I think is much much better suited to what the JGI needs, which is to have a protocol that’s really standardized, you know, that they can offer to their users.
ALISON: And JGI now does. Proposals have already started coming in. Some have been accepted, and more are being reviewed. If you’re interested in getting JGI support for your own isotope experiments, well, now you can. I’ll provide more info on how to apply in the show notes.
So, does Jennifer personally subscribe to the semi-automated technique?
JENNIFER: Oh, absolutely. Yeah, it’s all we use anymore.
ALISON: This episode was directed and produced by me, Alison Takemura, with editorial and technical assistance from Massie Ballon, Ashleigh Papp, and David Gilbert.
Genome Insider is a production of the Joint Genome Institute, a user facility of the US Department of Energy Office of Science. JGI is located at Lawrence Berkeley National Lab in beautiful Berkeley, California.
A huge thanks to Jennifer Pett-Ridge, senior staff scientist at Lawrence Livermore National Lab, in Livermore, CA. In addition to her work making stable isotope experiments much more doable, she leads Livermore Lab’s Environmental Isotope Systems Group. She also heads a U.S. Department of Energy Scientific Focus Area to understand mechanisms in the soil microbiome. [Side note: Jennifer’s team just ran their 1,000th SIP sample!]
You can hear more from Jennifer about the research questions that motivated her to collaborate with JGI on SIP automation. Just check out our website for a 10-minute recorded talk that we’ve posted to go along with this episode.
If you enjoyed Genome Insider and want to help others find us, leave us a review on Apple Podcasts, Google Podcasts, Spotify, or wherever you get your podcasts. If you have a question or want to give us feedback, Tweet us @JGI, or record a voice memo and email us at JGI dash comms at L-B-L.gov. That’s jgi dash c-o-m-m-s at l-b-l dot g-o-v.
Do you want to dive deeper into the research that JGI enables? If so, then join us at the JGI Annual Genomics of Energy & Environment Meeting, August 30th through September 1. It’s virtual and free and open to anyone who’s interested: from students to researchers to biology enthusiasts. Reserve your spot now by going to our website usermeeting.jgi.doe.gov.
And because we’re a user facility, if you’re interested in partnering with us, we want to hear from you! We have projects in genome sequencing, synthesis, transcriptomics, metabolomics, and natural products in plants, fungi, algae, and microorganisms. If you want to collaborate, let us know!
Find out more at jgi.doe.gov forward slash user dash programs.
If you’re interested in hearing about cutting edge research in secondary metabolites, also known as natural products, then check out JGI’s other podcast, Natural Prodcast. It’s hosted by Dan Udwary and me.
That’s it for now. See ya next time!
Additional information related to the episode
- Webinar: SIP technologies at the Environmental Molecular Sciences Laboratory (EMSL) and the JGI
- JGI’s Dariia Vyshenka uses SIP at the 2020 Berkeley Lab Research Slam in her flash talk: “Untangling Microbiomes”
- If you’re interested in doing SIP with the JGI, you can apply to do so through two of JGI’s user programs:
Simply submit a research proposal. Here’s more information on JGI’s Calls for User Proposals.