DOE user facilities EMSL and JGI announce FY 2019 collaborative FICUS projects.
Two Department of Energy user facilities, the Environmental Molecular Sciences Laboratory (EMSL) and the Joint Genome Institute (JGI), have selected 12 of the 41 proposals received from a joint call for 2019 research under the Facilities Integrating Collaborations for User Science (FICUS) initiative. This was the sixth FICUS call between EMSL and JGI since the collaborative science initiative was formed in 2014 by the Office of Biological and Environmental Research (BER) to harness the combined expertise and resources of two of the national user facilities stewarded by the DOE Office of Science in support of DOE’s energy, environment, and basic research missions.
Through the EMSL-JGI FICUS calls, users can combine EMSL’s unique imaging, omics and computational resources with cutting-edge genomics, DNA synthesis and complementary capabilities at JGI. Because the facilities offer a breadth of characterization and analytical power, researchers are not expected to be expert users on all of the scientific instruments. Instead, members of the scientific staff from both user facilities work with the researchers to assess their needs and help them with their methodologies, experiments and visualizations.
The accepted proposals began on October 1, 2018, and are:
-
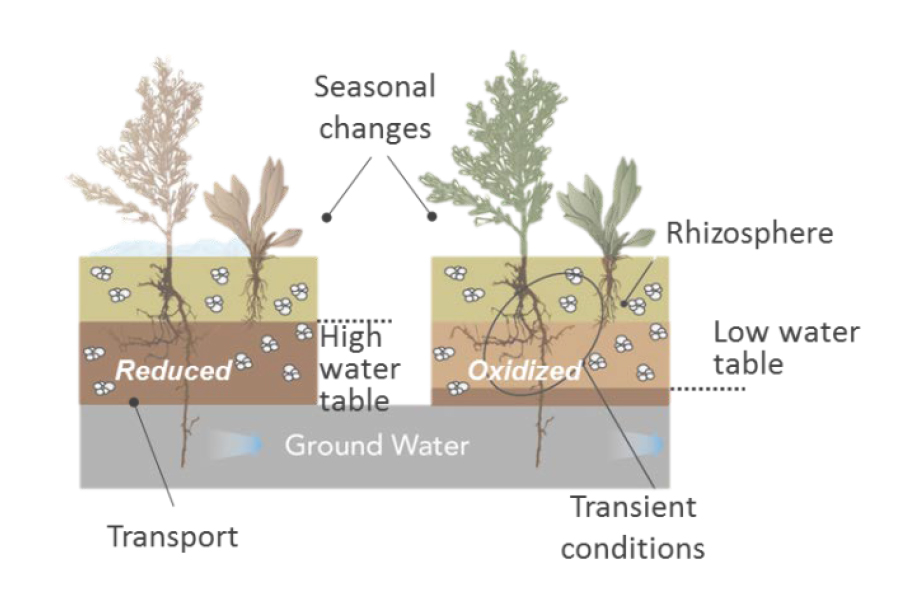
- Kristin Boye and Co-PI Christopher A. Francis of Stanford Linear Accelerator Center and Stanford University are examining the microbial metabolic activity and biogeochemical reaction networks in redox cycled alluvial systems. Redox-driven reaction switches are tied to water saturation and organic matter availability, and they regulate the retention, transformation, and release of organic carbon, nutrients, and contaminants that can impact groundwater quality. The study will focus on a uranium-contaminated floodplain near Riverton, WY, where significant seasonal fluctuations in redox conditions are indicated by temporary and spatially heterogeneous presence of sulfides, dissolved Fe and Mn (reducing conditions), and Fe oxides (oxidizing conditions). These fluctuations appear tied to water saturation and organic matter availability, and they drive the export of organic carbon, nutrients, and contaminants (U, Mo) to groundwater and surface water.
-
- Ronald Peter deVries and Co-PI Adrian Tsang (Co-PI) from Westerdijk Fungal Biodiversity Institute and Concordia University, respectively, are seeking to validate metabolic models of evolutionarily diverse fungi that were generated using an orthology-based approach to extrapolate from the reference sugar catabolism genetic network of Aspergillus niger. Knowledge of sugar catabolism is not only relevant in understanding the roles and abilities of these fungi in natural habitats, but can also be exploited to construct synthetic pathways for the production of valued and/or bioinspired chemicals and materials.
- Erik F. Y. Hom and Co-PI Jason E. Stajich from the University of Mississippi and University of California, Riverside, are probing microbial interactions and coordinated trophic responses in biological soil crusts (BSCs), which are the dominant ground cover in arid and semiarid drylands comprising ~40% of the global land surface. BSCs carry out important ecological services such as carbon and nitrogen fixation, the production of soil organic matter, soil water retention, redistribution of rainfall runoff, the provision of food sources for microflora and fauna, and the facilitation of vascular plant growth. This proposal focuses on understanding how BSC microbes interact–metabolically and physically–to impact one aspect of BSC ecosystem service and function: that of collective carbon and nitrogen cycling.
- James J. Moran from the Pacific Northwest National Laboratory, is looking to link phosphorus and carbon cycling in the rhizosphere. Root exudates provide a carbon source for rhizosphere microbial communities. In return, these communities help mobilize nutrients needed for plant growth. The team hypothesizes that spatial localization of root exudation maximizes the phosphorus return to the plant by leveraging dispersed microbial and geochemical microenvironments within the rhizosphere. They plan to study switchgrass microcosms in an effort to better understand the fundamental controls of nutrient delivery from soil to this bioenergy crop. Enhanced understanding of the linkages between carbon and phosphorus cycling in the rhizosphere will support switchgrass growth on marginal lands in support of DOE’s mission focus in bioenergy production.
- Michelle A. O’Malley of the University of California, Santa Barbara, is seeking to decipher the structure and function of secondary metabolites from anaerobic fungi. Building on recent discoveries of the biosynthetic machinery responsible for producing a wide array of secondary metabolites within the genomes of the anaerobic fungal phylum Neocallimastigomycota, O’Malley’s team hypothesizes that anaerobic gut fungi, during stressful conditions such as low nutrient availability, oxygen presence or microbial competition, synthesize secondary metabolites, enabling them to survive and thrive as important members of the consortia. O’Malley’s goal is to use this study to accelerate the identification of novel metabolites from anaerobic fungi, which could be used directly as biofuels and bioproducts or harnessed as tools to modulate microbial consortia.
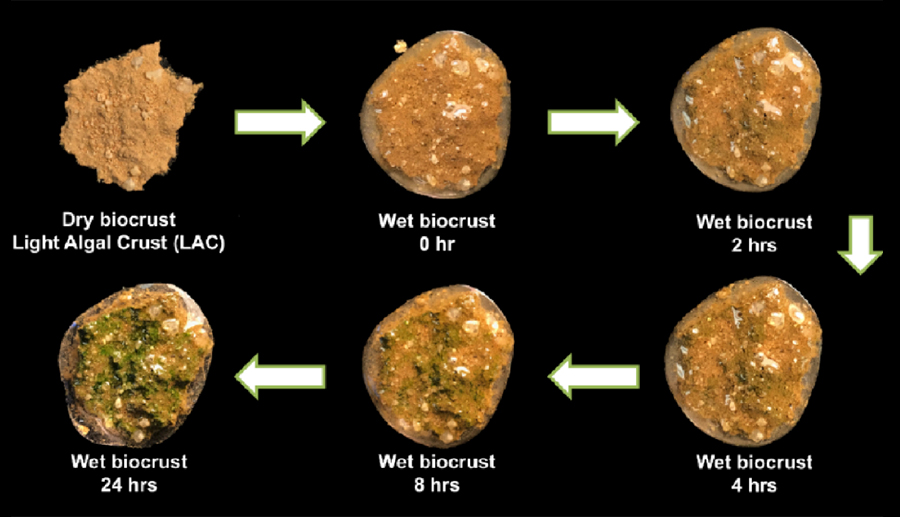
A light algal crust being reanimated with the addition of water, greening over the 24-hr time course. (Erik F. Y. Hom)
-
- Ryoko Oono from the University of California, Santa Barbara, is seeking to understand plant litter degradation and microbial defense by host-specific fungal endophytes, examining the transcriptomic, proteomic, and metabolomic specificity of diverse endophytic fungal species in response to their host litter and competing fungi. Litter senescence is an unexplored stage of rapid colonization and conversion of plant tissue matter by host-specific obligate endophytic fungi for fruiting body development. The results could transform the current paradigm of microbial community succession and function in litter and serve to explain a linkage between species diversity and major ecosystem processes, such as carbon cycling. Oono expects the insights from this study will enable coupling of the unexplored functions of diverse endophytic fungi to the production of biofuels, antibiotics, and novel enzymes that will be useful to human applications.
- Yeala Shaked and Co-PI Rhona Stuart from Hebrew University of Jerusalem and Lawrence Livermore National Laboratory, respectively, are examining the interactive mechanisms of mineral dissolution by microbial consortia. Essential metabolic functions within ecosystems are often partitioned among various members of microbial communities that each benefit the whole population. Such interdependencies structure microbial communities and can increase energy utilization efficiency and nutrient availability. Defining these interactions is critical for understanding the relationship between metabolic rates and elemental cycles in terrestrial and aquatic environments. In this study, Shaked and Stuart focus on the genetic and molecular basis of a symbiotic relationship between heterotrophic bacteria and a photosynthetic diazotroph, Trichodesmium, to understand the chemical pathways by which Trichodesmium/epibiont consortia solubilize and take up mineral- associated elements. Success will provide a genetic and molecular basis for mineral dissolution mechanisms that can be represented in ecosystem models of elemental cycling or engineered to improve bioenergy production.
- Kevin V. Solomon and Co-PI Scott D. Briggs from Purdue University are studying the impact of epigenetic regulation in anaerobic fungi to develop strategies that control lignocellulolytic enzyme expression and activity for increased lignocellulose breakdown and biofuel production. Solomon and Briggs seek to characterize the magnitude of epigenetic regulation globally on specific transcript and protein expression profiles in anaerobic fungi, and associate them with specific histone and DNA modifications at gene loci. They will then characterize local chromatin structure around these modifications to evaluate the mechanisms of regulation and identify promising epigenetic targets for further engineering.
- Cong T. Trinh from the University of Tennessee is seeking to understand and harness the robustness of undomesticated Yarrowia lipolytica strains for biosynthesis of designer bioesters. By leveraging genomics and molecular characterization at JGI and EMSL, Trinh plans to take a holistic approach to illuminate the underlying mechanisms of how lipolytica 1) tolerates and effectively assimilates inhibitory biomass hydrolysates for superior lipid accumulation under hypoxic conditions, 2) tolerates organic solvents beneficial for producing biofuels/bioproducts in a two-phase fermentation system, and 3) endogenously degrade lipids and produce esters with broad applications such as fuels, solvents, flavors, and fragrances. Success of the proposed research not only generates substantial fundamental knowledge for understanding and harnessing exceptional robustness of Y. lipolytica as a bioenergy microbial platform but also advances the microbial conversion technology to produce a diverse class of biofuels/bioproducts from lignocellulosic biomass.
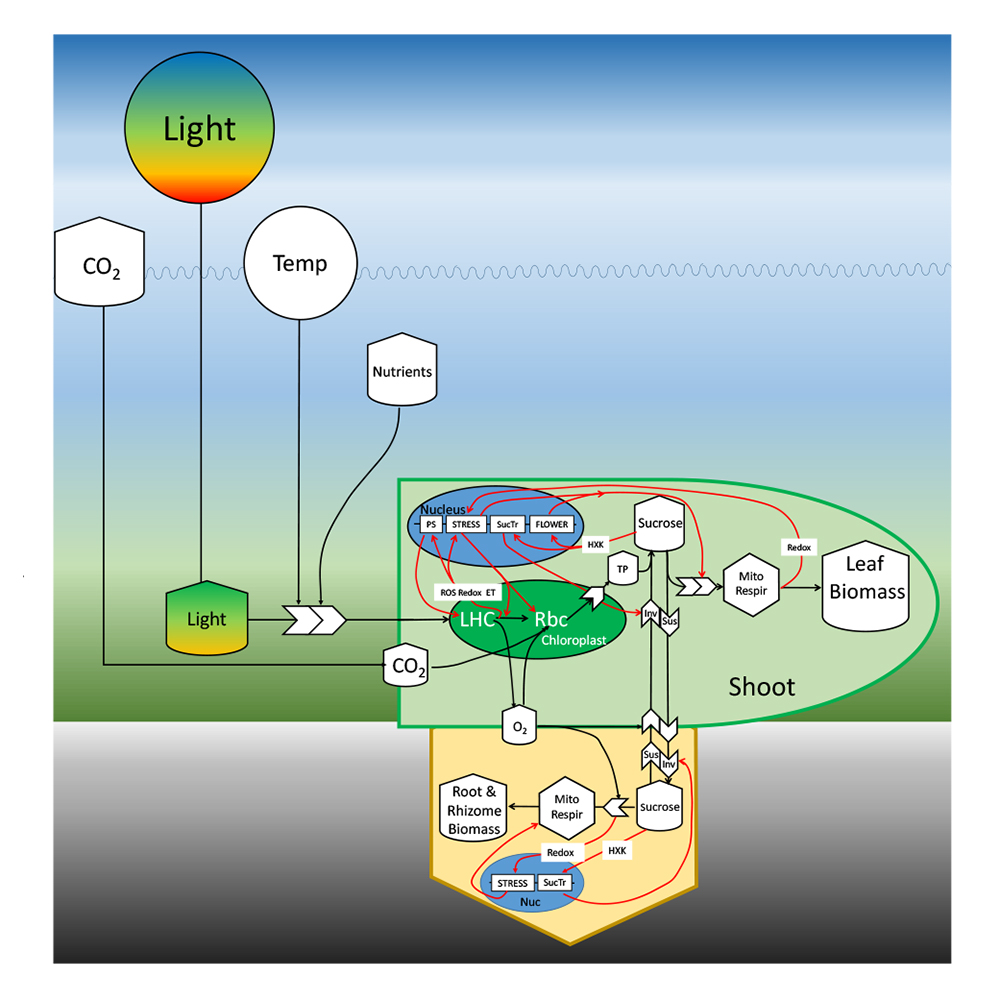
Schematic diagram illustrating the transcriptomic and metabolomic linkages between environmental drivers and eelgrass metabolic performance. (Richard Zimmerman)
- David J. Weston and Co-PI Wellington Muchero of Oak Ridge National Laboratory are examining the consequences of plant genetic variation and the surrounding microbiome on Sphagnum associated nitrogen fixation at elevated temperatures. Sphagnum has one of the largest impacts on global carbon fluxes of any single plant genus, and its productivity is largely due to the unique interactions with its associated microbiome and environment. The researchers will combine genomic and proteomic studies with isotopically labeled imaging measurements to understand nitrogen fixation and plant-microbe symbiosis. They are particularly interested in exploring their recent discovery that Sphagnum grows better at elevated temperatures when inoculated with a warming-adapted microbiome.
- Michael J. Wilkins of Colorado State University is seeking to understand climate and hydrologic feedbacks on hyporheic carbon processing in an upland watershed in western Colorado. The research will focus on processes occurring in the riverbed of a low-order fluvial system across seasonal time-points, where shifting patterns of river water – groundwater mixing have been observed that can potentially create strong redox gradients. The work will offer new insights into how river hydrology, carbon chemistry, and microbial activity shift over tractable time-scales. There is a strong need to better constrain these interactions and feedbacks, given that such mountainous upland catchments are experiencing rapid rates of ecosystem change linked to chronic environmental shifts. Results arising from this study will inform watershed-scale biogeochemical models to better predict carbon processing and export in upland ecosystems under a range of future environmental scenarios.
- Richard Carl Zimmerman from Old Dominion University is seeking to understand the physiological and genetic responses of eelgrass (Zostera marina L.) populations to the different temperature and CO2 environments that are predicted to alter the structure and function of coastal marine ecosystems in the next century. Eelgrass is the most widely distributed, and often dominant, seagrass species in the temperate northern hemisphere. The team plans to compare the differential responses of geographically isolated and genetically distinct populations from the seasonally dynamic thermal environment of the Chesapeake Bay, Virginia with those from stenothermal habitats in Puget Sound, Washington. This work will provide a holistic examination of how the environment influences critical downstream performance features linked to plant survival and contribute to predictive models identifying indicators of the overall health of seagrasses.
Click here for a full list of accepted projects under the JGI-EMSL FICUS initiative. Questions regarding the FICUS initiative may be directed to the EMSL User Support Office or the JGI Project Management Office.
* * *
EMSL, the Environmental Molecular Sciences Laboratory, is a DOE Office of Science User Facility that leads molecular-level discoveries that translate to predictive understanding and accelerated solutions for national energy and environmental challenges. EMSL supports the BER mission to achieve fundamental understanding and prediction of complex biological, Earth, and environmental systems for energy and infrastructure security.