DOE JGI researchers describe the current challenges in single-cell genomics.
The Science:
DOE JGI researchers review the status of single-cell genomics, and how close scientists are to being able to reconstruct an individual cell’s genome.
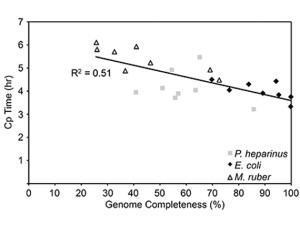
Graph of genome recovery vs. real-time MDA crossing point (CP) value from the results from a benchmark single-cell experiment using three reference strains with finished genomes: Pedobacter heparinus; Escherichia coli; and Meiothermus ruber. The median amount of genome recovered for the three bacterial strains ranged from 49 percent for M. ruber to 93 percent for E. coli. (Image from Clingenpeel et al., Front. Microbiol. doi: 10.3389/fmicb.2014.00771)
The Impact:
As an alternative method of studying microbial communities, single-cell genomics allows researchers to link function to phylogeny without needing to culture the microbes. Already, partial microbial genomes recovered from single cells are providing researchers with metabolic information and clues about population genetics.
Summary
As most of the microbes in, on, and around the plant are undiscovered or unculturable, single-cell genomics affords researchers a way to study microbial communities in their environment without having to culture them. The technology allows researchers to look at an individual microbe’s function within the community and improve the process of classifying microbial genetic relationships. Still, a single standard operating procedure for the process of recovering a single-cell genome from an environmental sample has yet to be developed.
Though researchers at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science user facility, have been able to successfully derive a complete genome from a single bacterial cell, this result remains the exception rather than the norm. In a perspective article published January 8, 2015 in Frontiers in Microbiology, a team of DOE JGI researchers led by Microbial Program head Tanja Woyke detailed the various challenges encountered, starting with the process of prepping the sample and isolating single cells for study. The process of isolating a single cell generally requires using fluorescence activated cell sorting (FACS), and researchers run across cells that are too big or too small, or too attached to other cells or particles.
Once single cells have been isolated and the DNA has been extracted and amplified for sequencing, it turns out the quality of the genome recovered can vary wildly. The team conducted an experiment, generating single cell genomes of three different, Gram-negative bacteria – Pedobacter heparinus DSM 2366, Escherichia coli K12-MG1655, and Meiothermus ruber DSM 1279 – for which the DOE JGI already had complete genome sequences. They found that the median amount of the genome recovered for the E. coli strain was 93 percent, but the M. ruber strain was only 49 percent. The researchers posited a number of reasons for the variation, from M. ruber being more resistant to the process of having its cell walls broken down to extract the DNA compared to the other bacteria, to limited genome access.
“For the time being, we will have to settle with partial genomes from a fraction of the cells contained within an environmental sample and reconstructing each cell’s genome from a complex environment still remains a dream rather than reality,” the researchers concluded. “However, these partial single-cell genomes are clearly pushing microbial genomics into exciting, untapped territory, enabling the discovery of unexpected metabolic features, providing insights into population genetics, and improving the phylogeny of microbes.”
Contact
Tanja Woyke
Microbial Program Head
DOE Joint Genome Institute
twoyke@lbl.gov
Funding
- U.S. Department of Energy Office of Science
Publication
Clingenpeel S et al. Reconstructing each cell’s genome within complex microbial communities – dream or reality? Front. Microbiol. 2015 Jan 8. doi: 10.3389/fmicb.2014.00771.