Data from marine heatwave event may foreshadow climate change impact on marine microbial communities.
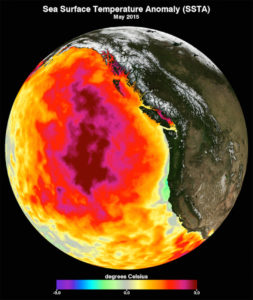
This data image shows the monthly average sea surface temperature for May 2015. Between 2013 and 2016, a large mass of unusually warm ocean water–nicknamed the blob–dominated the North Pacific, indicated here by red, pink, and yellow colors signifying temperatures as much as three degrees Celsius (five degrees Fahrenheit) higher than average. Data are from the NASA Multi-scale Ultra-high Resolution Sea Surface Temperature (MUR SST) Analysis product. (Courtesy NASA Physical Oceanography Distributed Active Archive Center)
The Science
A team of researchers tracked the impact of a large-scale heatwave event in the ocean known as “The Blob.” The event was recorded during a long-term study of microbial communities in the northeastern subarctic Pacific Ocean.
The Impact
The microbial communities in the ocean drive the biological pump that takes carbon from the atmosphere and keeps it in the deep ocean. With genomic samples collected before, during and after The Blob, researchers developed a preliminary model of how marine microbial communities are affected by warming events. More time-series studies could help researchers improve predictive models on how climate change may impact marine microbes and by extension, the ocean’s capacity to act as a carbon sink.
Summary
As global temperatures rise, so do instances of extreme weather events such as hurricanes and droughts. Another one began in late 2013, when a large patch of warm ocean water about a third the size of the United States began forming in Alaska. Over the next two years it slowly spread, raising water temperatures by a few degrees. Eventually The Blob reached southern California.
Steven Hallam of the University of British Columbia was already collecting ocean water samples when The Blob started. The samples were taken up and down the water column along the Line P transect, a series of geographic coordinates which go from coastal to open ocean waters.

CTD Rosette in use along the Line P transect. A CTD is used to profile a water column from surface to bottom by measuring conductivity (a proxy for salinity), temperature, and depth. (Jody Wright)
Between 2010 and 2016, Hallam and his colleagues collected more than 270 water samples. Genomic DNA extracted from the water samples were sequenced by the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), as part of Hallam’s approved proposal through the Community Science Program. The proposal focuses on uncovering the metabolic networks involved in carbon and nutrient flux in marine microbial communities.
Using the JGI-generated data, Sachia Traving, a former postdoctoral fellow in the Hallam lab now at the University of Southern Denmark, and colleagues at Hakai Institute, and Fisheries and Oceans Canada’s Institute of Ocean Sciences modeled the impact of The Blob on the microbial communities up and down the water column. They recently shared their findings in Communications Biology. They reported that the heatwave caused the microbial populations to cluster along the water column, like layers in a cake. Additionally, as nutrients became scarce during the heatwave, they saw populations shift. Smaller picoplankton and free-living microbes that can fix carbon dioxide replaced organic matter scavengers. This preliminary model aligns with several other studies on the potential effects of warming oceans on the microbial populations and the global carbon cycle.
Tracking large-scale marine events such as The Blob is difficult, but critical. By 2016, the microbial populations had begun to recover from the heatwave. Hallam said the work underscores the value of large-scale research collaborations and of conducting even more time-series studies.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
PI Contact
Steven Hallam
University of British Columbia
shallam@mail.ubc.ca
Funding:
This work was performed under the auspices of the Scientific Committee on Oceanographic Research (SCOR), the US Department of Energy (DOE) Joint Genome Institute, an Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02- 05CH11231, the G. Unger Vetlesen and Ambrose Monell Foundations, the Natural Sciences and Engineering Research Council of Canada, the Canada Foundation for Innovation, and the Canadian Institute for Advanced Research through grants awarded to S.J.H. S.J.T. was supported by the Danish Research Council (grant DFF-7027-00043B) and CTEK by the Tula Foundation.
Publication:
- Traving SJ et al. Prokaryotic responses to a warm temperature anomaly in northeast subarctic Pacific waters. Commun Biol. 2021 Oct 22;4(1):1217. doi: 10.1038/s42003-021-02731-9.
Related Links:
- University of British Columbia story: “Heatwaves like ‘the Blob’ could decrease role of ocean as carbon sink”
- Hallam Lab Line P Project
- JGI Community Science Program
Byline: Massie S. Ballon