The success of an RNA project is primarily dependent on the quality of the material received from the collaborator. Samples not meeting minimum submission requirements with regard to quantity and quality will result in a delay to project initiation. If you have any questions or concerns, contact your Project Manager or the Project Management Office.
View protocols for isolating RNA, provided by the JGI user community. View the sample QC protocols that JGI will use to check the quality of your RNA.
RNA sample requirements:
The JGI uses total RNA as starting material for all of its library protocols.
Different types of samples may require specific RNA isolation techniques. JGI has successfully used standard methods involving Trizol, Rneasy and for many samples further purified using LiCl. Contaminants in the preparation will often cause an overestimate of RNA quantity by OD measurement. It is advisable to submit RNA of the highest quality possible since degraded/fragmented RNA will introduce noise into the sequencing data.
All samples must meet the following criteria:
- All samples must be treated with RNase-free DNase prior to submission. Contaminating DNA makes an ideal template for cDNA library construction, and reads originating from DNA can’t be informatically separated from reads originating from RNA, so samples must be DNase-treated. Many kits are available; the Invitrogen Turbo DNA-free kit removes DNA without columns or precipitation (please make sure to do the DNase inactivation step). For samples that will be used for rRNA depleted libraries, we recommend using this kit after RNA extraction even if the extraction protocol includes a DNase treatment step.
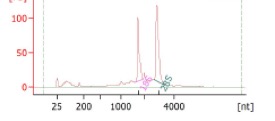
- RNA must not show signs of degradation as measured by distinct bands of ribosomal peaks of relative intensities (gel electrophoresis or Bioanalyzer results). Please see example at the end of the document. The JGI does not require submission of either a gel photo or Bioanalyzer/Fragment Analyzer trace with samples. However, Investigators are strongly advised to assess the quality of materials to be submitted by gel electrophoresis (including a standard molecular weight marker) or by electropherograms such as Bioanalyzer or Fragment Analyzer.
- A260/280 greater than 1.8 (spectrophotometer/NanoDrop). A lower ratio is often indicative of protein/DNA contamination; a ratio higher than 2.1 may indicate residual guanidine thiocyanate or beta-mercaptoethanol. Protein contaminants should be re-extracted using phenol:chloroform:isoamyl alcohol; other contaminants by EtOH ppt. In general, JGI recommends LiCl extraction for cleaning up RNA.
- Quantification must be performed by fluorometric measurement (for example, Qubit Fluorometer – Life Technologies, Inc.). Samples that are quantified using a spectrophotometer or NanoDrop will not be accepted.
The table below should be used as a guide for the amount of total RNA required for the most common JGI library types:
| Library Type | Selection method | Requested mass (ng) | Tube volume (ul) | Plate volume (ul) | Concentration range (ng/ul) |
| Eukaryotic RNA | polyA selection | 3000 | 25-500 | 25-150 | 50-1000 |
| Eukaryotic RNA – low input | polyA selection | 300 | 25-500 | 25-150 | 10-1000 |
| Eukaryotic smRNA | 3000 | 25-500 | – | 166-1000 | |
| Eukaryotic Iso-Seq |
1000 | 25-500 | – | 80-1000 | |
| Prokaryotic RNA – low input | rRNA depletion | 300 | 25-500 | 25-150 | 10-1000 |
| Prokaryotic smRNA | 13000 | 25-500 | – | 166-1000 |
The actual amount of total RNA needed may vary by individual project. Please consult with your Project Manager if you may not be able to provide adequate material.
- For small RNA, extraction kits are commercially available for isolation of total RNA containing miRNA or small RNA (<200bp). Please refer to the manufacturer’s instructions to ensure the protocol will retain RNA <200 bp.
Shipping:
Prior to shipping samples to the JGI, all sample metatdata must be completed in its entirety. If you have questions about required information, consult with your Project Manager early in the submission process.
- RNA samples are most stable in RNase-free water. RNA should be completely dissolved and shipped on dry ice.
- Individual RNA samples should be shipped to the JGI in a single tube (or well of a plate). Preps suitable for pooling should be pooled prior to shipping to the JGI. You must ship all samples in the barcoded tubes or plates provided by the JGI.
- Do not ship samples until you have received a shipping approval email from the JGI. Samples that have not been approved will be returned to sender.
Example of Bioanalyzer results of RNA:
Total RNA: Clear 18s and 28s rRNA peaks; no degradation.
Last update: 4/20/23