Metagenomics and single cell strategies help reveal a novel bacterial phylum.
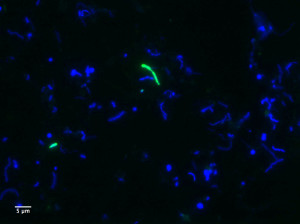
A ‘Ca. Kryptonia’-specific FISH probe was designed and used to visualize cells from Dewar Creek Spring sediment samples. ‘Ca. Kryptonia’ cells hybridizing with the probe are green, while other cells are visualized with 4′,6-diamidino-2-phenylindole (DAPI; blue). (Composite image by Emiley Eloe-Fadrosh, DOE JGI)
Although global microbial populations are orders of magnitude larger than nearly any other population in, on or around the planet, only a fraction has been identified thus far. The U.S. Department of Energy (DOE) is seeking to uncover the true extent of the planet’s microbial diversity in order to learn more about the genes, enzymes and metabolic pathways that play key roles in regulating critical biogeochemical cycles. More thorough surveys could lead to new strategies for DOE researchers to advance their energy and environmental investigations.
In a study published January 27, 2016 in Nature Communications, a team led by researchers at the DOE Joint Genome Institute (JGI), a DOE Office of Science User Facility, utilized the largest collection of metagenomic datasets to uncover a completely novel bacterial phylum that they have dubbed “Kryptonia.”
“We were interested in looking for novel, divergent bacterial or archaeal sequences that hadn’t been previously characterized,” said study first author Emiley Eloe-Fadrosh, a DOE JGI research scientist. “We didn’t have a particular target to go after, but reasoned that there was likely a wealth of untapped diversity just waiting to be discovered in all the metagenomic data.”
A Signal in the Noise

Watch Emiley Eloe-Fadrosh on the search for Kryptonia from the DOE JGI’s 2015 Genomics of Energy and Environment Meeting at http://bit.ly/JGI15UMKryptonia.
A researcher analyzing vast quantities of genomic data is not unlike a beachcomber slowly scanning a beach with a metal detector. Both are searching for a signal in the noise that indicates buried treasure, be it novel microbes or pirate gold. The team started with 5.2 trillion bases (Terabases or Tb) of sequence in the Integrated Microbial Genomes with Microbiome Samples (IMG/M) system. After scouring this equivalent of over 1,700 human genomes or 1 million E. coli bacterial genomes the team identified long sequences that contained a phylogenetic marker (DNA corresponding to ribosomal RNA, rRNA) commonly used to assign all life (bacteria, archaea, and eukaryotes) into a particular classification system.
The team identified sequences from four different geothermal springs – Great Boiling Spring, Nevada, Dewar Creek Spring in Canada, and the Gongxiaoshe and Jinze pools in China – that could not be placed into any recognizable phylum. Reconstructing the genomes from metagenomic datasets and single cell genomes yielded four lineages belonging to the novel candidate phylum, named Kryptonia (Candidatus Kryptonia) from the Greek word for “hidden.”

Among the hot springs where Kryptonia was found is Gongxiaoshe Spring in China, shown with the rural Chinese village of Dientan in the background. (Brian Hedlund, UNLV)
Given that there are currently 35 cultured bacterial and archaeal phyla, and roughly the same number of recognized uncultured phyla, Eloe-Fadrosh says the identification of a novel candidate phylum was a surprise. “It’s not every day that you find a completely new phylum. With all the studies that have been conducted in hot springs, there’s an assumption that all novelty has been found. But we found these unknown lineages in high abundance.” UPDATE: A complementary paper in Nature Microbiology speculated on the microbial lineages that remain unknown due to technology biases. Read more about this at http://jgi.doe.gov/strategy-to-uncover-more-microbial-lineages/.
The analysis of the nearly complete Kryptonia genomes recovered from this metagenomics dataset also revealed the presence of the CRISPR-Cas phage defense system in these organisms. Using this information, the team was able to track the global biogeographic distribution of Kryptonia vis-à-vis the putative phages infecting the bacteria. “While a lot of research and media attention is gathering around the biotechnological applications of the CRISPR-Cas system, we are very excited about using it as a powerful tool in reconstructing the infection history of the organisms, as well as a fingerprint to uncover and trace the correlated viruses,” said Prokaryote Super Program Head Nikos Kyrpides, a co-author of the paper.
Uncovering Novelty Also Reveals Biases
Analyses of Kryptonia reveal that the bacteria need to rely on other microbes for several nutritional requirements, suggesting a reason this candidate phylum had not been found previously despite its abundance in geothermal springs. “We hypothesize that Kryptonia engages in a metabolic partnership, and it’s very challenging to cultivate bacteria that have unique interactions in the wild that can’t necessarily be replicated in the lab,” said Eloe-Fadrosh, who credited the combined power of metagenomics and single-cell genomics to capture the novel microbes described in the paper. “I think one of the grand challenges for the field is to quantify microbial diversity, and these technologies are getting us closer to making that a reality.”
The work reinforces the perspective published in Science last year by DOE JGI Director Eddy Rubin and Microbial Program head Tanja Woyke. “There are reasons to believe that current approaches may indeed miss taxa, particularly if they are very different from those that have so far been characterized,” they wrote. “Past explorations of available metagenomic data sets have focused on the discovery of matches to the known genes and genomes—an analysis that is naturally biased against uncovering completely novel life.”

Sequences from Great Boiling Spring in Nevada have been attributed to the novel candidate phylum Kryptonia. (Brian Hedlund, UNLV)
Eloe-Fadrosh said the team found unique metabolic pathways in Kryptonia, hints that there may be other novel enzymes related to biological pathways waiting to be uncovered. “Just like Taq polymerase was revolutionary to molecular biology, there could be an enzyme in Kryptonia with biotechnological relevance,” she added.
Her words echo the speculations offered by study co-author and DOE JGI collaborator Brian Hedlund of the University of Nevada, Las Vegas. Noting that Kryptonia play a role in lignocellulose degradation, he added that there are potential resources still waiting to be tapped for biotechnology applications in the “microbial dark matter” from which Kryptonia has only just emerged. For example, he said, companies are marketing enzymes from thermophiles for quick diagnostic tests including those from previous research enabled by the DOE JGI on Yellowstone hot pools. “I do believe that applications are there if people spend time and money looking for the microbes,” he said.
Eloe-Fadrosh spoke about the search for Kryptonia at the DOE JGI’s 2015 Genomics of Energy and Environment Meeting. Watch the video at http://bit.ly/JGI15UMKryptonia.
Aside from Hedlund at the University of Nevada, Las Vegas, collaborators on this work include researchers at: University of Calgary (Canada); Lawrence Livermore National Laboratory; Geological Survey of Canada; McMaster University (Canada); Miami University; University of Alaska-Anchorage and Sun Yat-Sen University (China).