Researchers build a genetic profile for a section of Aspergillus fungi.
There are millions of fungal species, and those few hundred found in the Aspergillus genus play important roles in areas ranging from industrial production to agricultural plant pathogens. Reported October 22, 2018, in Nature Genetics, a team led by scientists at the Technical University of Denmark, the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility, and the Joint Bioenergy Institute (JBEI), a DOE Bioenergy Research Center, present the first large analysis of an Aspergillus fungal subgroup, section Nigri.
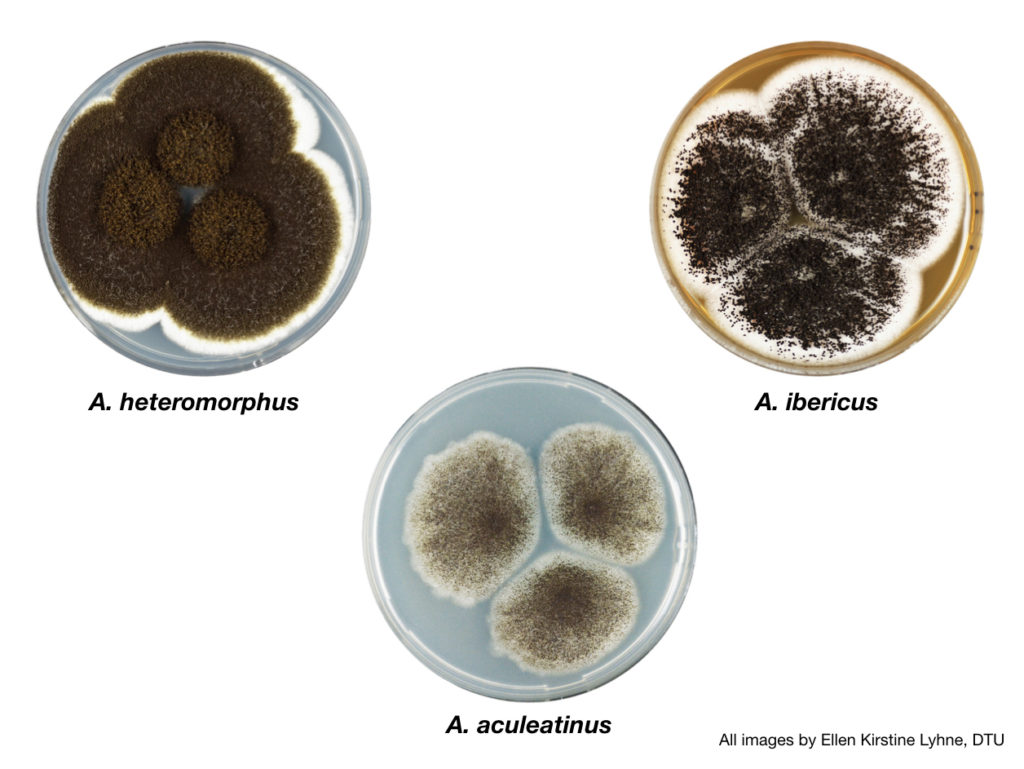
Aspergillus section Nigri fungi sequenced and analyzed for this study (clockwise from top left): A. heteromorphus; A. ibericus; and A. aculeatinus. (All images by Ellen Kirstine Lyhne, DTU.)
“Aspergillus is such an important genus, not just for biotechnology, but for all the roles it plays in agriculture, human health, enzyme production and food fermentation,” said study co-author Scott Baker, Science Area Lead at the Environmental Molecular Sciences Laboratory, a national user facility at Pacific Northwest National Laboratory and a member of JBEI’s Fungal Biotechnology group. “It’s the Swiss Army knife of fungal genera as far as its impact on things that are important to us.”
The results are part of a long-term project to sequence, annotate and analyze the genomes of more than 300 Aspergillus fungi. Six fungal genomes within section Nigri had already been sequenced previously; JGI sequenced and annotated 23 additional fungal genomes within the subgroup. These included genomes of 20 species and multiple isolates of the same species, Aspergillus niger. The resulting compendium allows researchers to conduct intraspecies comparisons within section Nigri, as well as compare the fungal genomes within the subgroup to other genomes within the Aspergillus genus.
Studying Species Diversification
“Comparison within and across a taxonomic section allows us to build a genetic profile for that section, which can be connected to the phenotypes (known characteristics) of the species in the section,” said study senior author Mikael Andersen of the Technical University of Denmark. “This is quite useful in many regards; for instance, to identify new unique enzymes or to link an interesting bioactive compound to the biosynthetic genes. This paves the way for new sustainable cell factories.”
Baker added that the intraspecies comparisons within the subgroup offer the researchers an opportunity to study species diversification and delve into evolution of fungi. “This gives us insight in the differences between these really closely related species,” he said. “When we engineer organisms, we’re making changes. So are there insights we can gain from the natural evolution process to help us better engineer strains that can do good things for us?”
In choosing to focus on section Nigri, the researchers recognized the myriad of industrial applications for the fungi within this subgroup. They serve as production organisms for 49 of 260 industrial enzymes and are also very efficient producers of enzymes and secondary metabolites. JGI Fungal Program head Igor Grigoriev said that one of the aims in sequencing the genomes within the Aspergillus genus is to increase the catalog of carbohydrate active enzymes (CAzymes), which can degrade plant cell walls and thus have applications in the DOE mission to develop industrial processes for producing sustainable alternative fuels using candidate bioenergy feedstock crops.
Thousands of New Genes
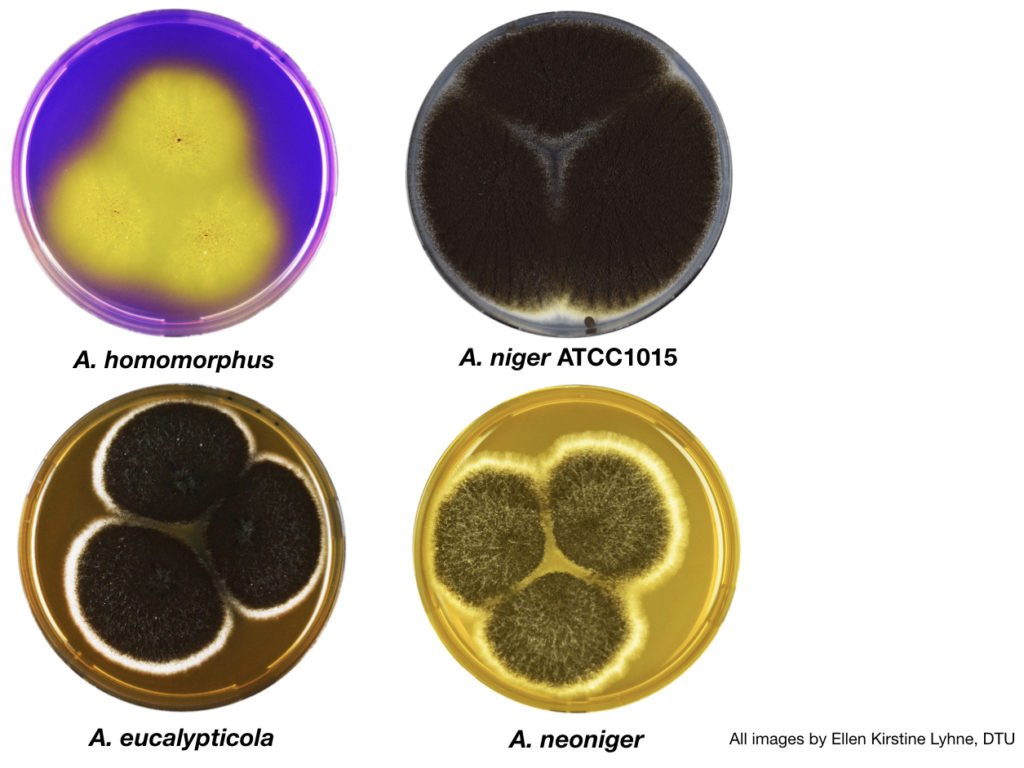
Aspergillus section Nigri fungi sequenced and analyzed for this study (clockwise from top left): A. homomorphus; A. niger ATCC1015; A. neoniger; and A. eucalypticola. (All images by Ellen Kirstine Lyhne, DTU.)
The team discovered thousands of new genes. For all sets of genes unique to section Nigri, they identified species-specific gene clusters that are related to functions such as secondary metabolism, gene regulation or protein regulation. Additionally, though there are shared gene clusters for secondary metabolite biosynthesis, all of the Aspergillus section Nigri fungi have their own unique sets of genes for secondary metabolism.
Andersen noted that as the team continues to sequence and annotate more genomes within the Aspergillus genus, they aim to report on more sections. “Many more interesting discoveries are on the way; the diversity is enormous, and the potential for bioenergy, biotechnology, and biotherapeutics in each and every species is staggering.”
All of the published Aspergillus genomes are publicly available from the JGI fungal genomes portal Mycocosm at genome.jgi.doe.gov/Aspergillus.
Publication: Vesth, TC et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nature Genetics. 22 October 2018. doi: 10.1038/s41588-018-0246-1