For years, Elizabeth (Toby) Kellogg and other researchers at the Danforth Plant Science Center (Danforth Center) drove up and down the highways of the continental United States, occasionally pulling over to the side of the road to collect small weedy plants and bring them back to the lab. The weedy grass was green millet (Setaria viridis), a small model grass with a short lifecycle that uses a carbon fixation process known as the C4 pathway, which particularly helps plants thrive in warm, arid environments. Corn and sugarcane are among the major high-yield C4 crops, as are the candidate biofuel feedstocks Miscanthus and switchgrass.
Innumerable road trips and hundreds of plants have resulted in a paper published October 5, 2020, in Nature Biotechnology. Kellogg and her colleagues, along with researchers at the HudsonAlpha Institute for Biotechnology and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), generated genome sequences for nearly 600 green millet plants and released a very high quality reference S. viridis genome sequence. Analysis of these plant genome sequences also led researchers to identify a gene related to seed dispersal in wild populations for the first time.
“To our knowledge, nobody has ever discovered a dispersal gene that way,” said Kellogg, a senior author of the paper. “This paper is the first one to survey a huge amount of natural diversity and say, ‘Yeah, there are genes out there that affect this phenotype.’”
Results from A “Massive Amount” of Sequencing

Green millet plants growing at the Danforth Center. (Bruton Stroube/ Donald Danforth Plant Science Center)
Seed dispersal is critical for plants in the wild, but it is an undesirable trait for domesticated crops because it leads to reduced harvest yields. Over thousands of years, farmers have selected for cereal plants without this shattering trait – referring to the moment when the cluster of seeds at the tip of each branch breaks apart so the seeds can disperse – so that the seeds remain atop the plant to be collected.
Association mapping led the team to identify a gene called Less Shattering 1 (SvLes1); gene editing studies led by co-first author Pu Huang confirmed that it was involved in shattering by turning it off. “It’s a new shattering gene variant identified in a natural population. Not very many of these shattering genes have been discovered that let a plant go all the way to seed but prevent the seeds from falling,” said JGI Plant Program head Jeremy Schmutz, who is also a HudsonAlpha Faculty Investigator. “This could be another mechanism to turn off shattering and domesticate crops.” How shattering occurs varies widely between crops, Kellogg added, and shattering genes may be specific to species or groups of species.
The genome data also revealed that green millet was introduced into the United States multiple times from Eurasia. The team also identified a gene associated with leaf angle, which determines how much sunlight leaves can get and in turn serves as a predictor of yield. The gene is an ortholog of known genes, “The gene has now been mapped back in maize as involved in leaf angle,” noted Schmutz. “It’s a nice example of de novo discovery and then mapping back to identify candidate genes.”
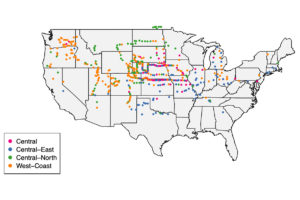
Every dot on this map represents the location of a green millet (S. viridis) plant collected by Danforth Center researchers. (Courtesy of the Kellogg lab)
Through JGI’s Community Science Program, sequences of several hundred green millet plant genomes were generated, though the final analyses focused on 598 individuals. Schmutz and his team assembled and annotated the genomes at HudsonAlpha. Sujan Mamidi and Adam Healey, two of the co-first authors, led the data analyses and assembled the green millet “pan-genome” (a set of 51,000 genes that represent all the genes that are present in a given species).
“This is a great example of developing a large-scale genome infrastructure with a reasonably accessible system,” said Schmutz. “Building the pan-genome and accessions allow us to see presence/absence variation easily and to find genes missing in particular accessions, and to confirm phenotypes, which validate traits.”
“The number of lines sequenced is not trivial, and they were all assembled de novo, which let the team look at presence/absence of whole genes,” Kellogg agreed. “Getting that information is hard. There’s a good reason nobody’s done it; it’s a heck of a lot of work. I wouldn’t have done it without the contribution of Jeremy’s group. It’s just a massive amount of sequencing.”
A Resource for Many Applications
Kellogg noted that C4 crops have gotten a lot of interest because they’re very productive even in high heat while C3 crops have become less efficient at photosynthesis, a concern as extreme weather events become more frequent. “A big part of the Danforth Center’s mission is to feed the hungry and improve human health. So there’s a major question: how to turn a C3 crop into a C4 crop. There should be a master regulator but no one has found it,” Kellogg mused. “[The S. viridis genome] is a resource for many different applications. The JGI group has been wonderful to collaborate with, and this [project] wouldn’t have been possible without their involvement; it’s something we wouldn’t have even started.”
The reference genome of Setaria viridis is available on JGI’s plant portal Phytozome.
Researchers from the RIKEN Center for Sustainable Resource Science (Japan) and Chinese Academy of Agricultural Sciences (China) were also involved in this work.
Publication: Mamidi S et al. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nature Biotechnology. 2020 Oct 5. doi:10.1038/s41587-020-0681-2
Byline: Massie Santos Ballon
***
About The Donald Danforth Plant Science Center: Founded in 1998, the Donald Danforth Plant Science Center is a not-for-profit research institute with a mission to improve the human condition through plant science. Research, education and outreach aim to have impact at the nexus of food security and the environment, and position the St. Louis region as a world center for plant science. The Center’s work is funded through competitive grants from many sources, including the National Institutes of Health, U.S. Department of Energy, National Science Foundation, and the Bill & Melinda Gates Foundation. Follow us on Twitter at @DanforthCenter.
About HudsonAlpha: HudsonAlpha Institute for Biotechnology is a nonprofit institute dedicated to developing and applying scientific advances to health, agriculture, learning, and commercialization. Opened in 2008, HudsonAlpha’s vision is to leverage the synergy between discovery, education, medicine, and economic development in genomic sciences to improve the human condition around the globe. The HudsonAlpha biotechnology campus consists of 152 acres nestled within Cummings Research Park, the nation’s second largest research park. The state-of-the-art facilities co-locate nonprofit scientific researchers with entrepreneurs and educators. HudsonAlpha has become a national and international leader in genetics and genomics research and biotech education and fosters more than 40 diverse biotech companies on campus. To learn more about HudsonAlpha, visit hudsonalpha.org.
