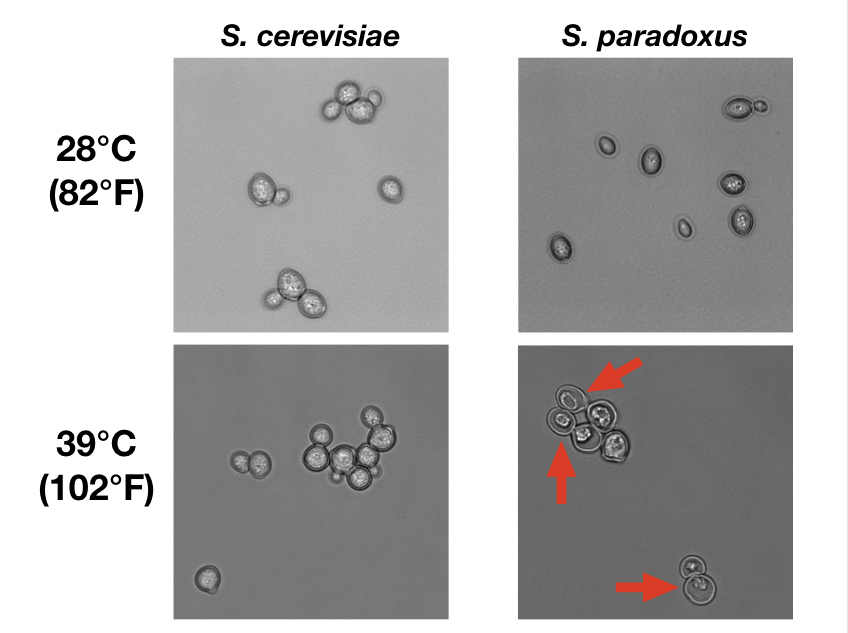
At high temperature, S. paradoxus cells die in the act of cell division, as seen by the dyads with cell bodies shriveled away – highlighted by arrows – from the outer cell wall. (Images by Carly Weiss, courtesy of the Brem Lab)
A new approach for improving functional annotation in fungal genomes.
The Science
In a proof-of-concept study, researchers demonstrated that a new genetic mapping strategy called RH-Seq can identify genes that promote heat resistance in the brewer’s/baker’s yeast Saccharomyces cerevisiae, allowing this species to grow better than its closest relative S. paradoxus at high temperatures (39°C/102°F).
The Impact
One of the challenges in understanding wild populations is that tests to find associations between genotype – an organism’s genetic makeup – and phenotype – an organism’s physical characteristics, properties and behaviors – can only be applied to interfertile members of a given species. These approaches don’t allow researchers to compare across very distant species that no longer mate to produce progeny. The ability to pinpoint which genes underlie unique traits in far-flung species would speed up efforts to harness species diversity in traits relevant to U.S. Department of Energy (DOE) missions in energy and environment.
Summary
There are more than 1,000 species of yeasts and they vary in terms of genetics, metabolism and biochemistry. Their capabilities can be harnessed for a wide range of biotechnological applications, including biofuel production. Efforts such as the 1000 Fungal Genomics Project led by the DOE Joint Genome Institute (JGI), a DOE Office of Science User Facility of Lawrence Berkeley National Laboratory, aim to improve our understanding of fungal diversity by sequencing genomes from across the fungal Tree of Life, but they also highlight the corresponding need to be able to assign functions to unknown genes and improve functional annotation to harness the genomic data.
Reported October 8, 2018, in Nature Genetics, a team led by researchers at the University of California, Berkeley and the Buck Institute for Research on Aging have demonstrated a way to map associations between genotype and phenotype using a strategy called reciprocal hemizygosity analysis via sequencing (RH-Seq). Strains of S. cerevisiae and S. paradoxus were first used to create a sterile interspecific hybrid. In this background, the researchers then used a series of random insertions of a foreign DNA sequence to generate a huge pool of mutants, each containing only a single copy of a given gene rather than the two that are present in the native state. During culture at 39°C, some of these half-dosage mutants died and others grew well; the survivors had their DNA sequenced at the JGI as part of a 2014 Community Science Program project. The results revealed that at each of eight genes, having a single version from S. cerevisiae was sufficient to promote growth at high temperature, and having the version from S. paradoxus led to very poor heat tolerance. This represents a first-ever case of complex genotype-to-phenotype mapping between species that have been separated for millions of years. The study highlights the genes that could be used to make other fungal species besides S. cerevisiae more thermotolerant, and it paves the way for dissection of additional traits, in fungi and beyond.
Contacts:
BER Contacts
Daniel Drell, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Rachel Brem
Buck Institute for Research on Aging
[email protected]
Funding:
The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under contract no. DE-AC02-05CH11231. The work was supported by R01 GM120430-A1.
Publication
- Weiss CV et al. Genetic dissection of interspecific differences in yeast thermotolerance. Nature Genet. 2018 October 8. doi: 10.1038/s41588-018-0243-4.
Related Links:
- JGI Community Science Program (CSP): https://jgi.doe.gov/user-program-info/community-science-program/
- Approved CSP FY14 proposal: Pioneering fungal mutagenesis using Tn-seq
- JGI News Release: Expanding the Stable of Workhorse Yeasts
- JGI Blog: Cracking the Code: Evolutionary Changes in the Genetic Code of Yeasts