New genome sequences target next generation of yeasts with improved biotech uses
The yeast Saccharomyces cerevisiae was a part of human civilization before history was recorded. It is essential for making bread, beer and wine, and it is ubiquitous. It is not, however, typical of the more than 1,500 yeast species found around the world. Yeasts are physically hard to distinguish, and it is easy to think they are all the same. Metabolically, genetically and biochemically, however, yeasts are highly diverse. Characteristics such as their thick cell walls and tolerance of pressure changes that could rupture other cells mean yeasts are easily scaled up for industrial processes. In addition, they are easy to grow and modify and, with notable exceptions such as Candida albicans, most are not associated with human illness. While these capabilities can be used for a wide range of biotechnological applications, including biofuel production, so far industry has only harnessed a fraction of the diversity available among yeast species.
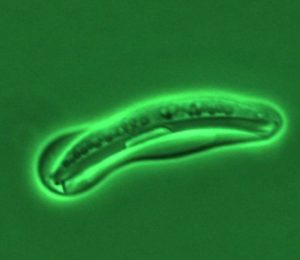
One of the genomes sequenced for this study was of the yeast Scheffersomyces stipitis. (Courtesy of Tom Jeffries, University of Wisconsin, Madison)
To help boost the use of a wider range of yeasts and to explore the use of genes and pathways encoded in their genomes, a team led by researchers at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility at Lawrence Berkeley National Laboratory, conducted a comparative genomic analysis of 29 yeasts, including 16 whose genomes were newly sequenced and annotated. In the study published the week of August 15, 2016 in the Proceedings of the National Academy of Sciences (PNAS), the team mapped various metabolic pathways to yeast growth profiles.
“Obtaining a complete genome of a microbe that is industrially important greatly stimulates research in the area,” said senior author Tom Jeffries, Professor Emeritus at University of Wisconsin, Madison, of the work. “This is particularly true when the genomic sequence is accompanied by a high-quality annotation of the genes, and the JGI annotation pipeline is one of the best in the field. We can expect an explosive interest in yeast biology in the coming years.”
Yeast genetic diversity wider than expected
Yeasts (which belong to the “kingdom” of fungi) can use a wide range of carbon and energy sources, ranging from cellulosic (6-carbon) and hemicellulosic (5-carbon) sugars to methanol, glycerol, and acetic acid. Products include ethanol and other alcohols, esters, organic acids, carotenoids, lipids, and vitamins. In fact vitamin complexes and some nutritional supplements are derived from yeasts.
“We sequenced these diverse genomes to expand the catalog of genes, enzymes, and pathways encoded in these genomes for producing biofuels and bio-based products we use in daily life,” said Igor Grigoriev, JGI Fungal Program Head and co-senior author of the manuscript.
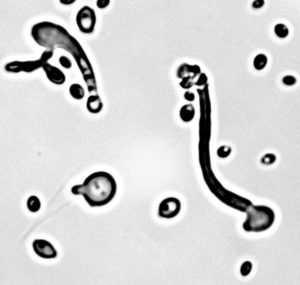
Researchers found a non-stop codon reassignment when they sequenced the genome of the yeast Pachysolen tannophilus. (Courtesy of Cletus Kurtzman, USDA-ARS)
Sequencing these less-known yeasts and characterizing their metabolic pathways, added study first author Robert Riley of the DOE JGI, helps fill in knowledge gaps regarding the fungal enzymes that can help convert a wide range of sugars into biofuel. The well-known yeast S. cerevisiae, for example, ferments glucose, but not the full range of sugars found in plant biopolymers.
One of the newly-sequenced yeasts is Pachysolus tannophilus, which can ferment xylose, otherwise known as wood sugar as it is derived from hemicellulose, which along with cellulose, is one of the main constituents of woody biomass. It is only distantly related to well-studied xylose fermenters such as Scheffersomyces stipitis — another yeast sequenced by the DOE JGI.
These distances are huge. “We might think of yeasts as simple unicellular, creatures similar to each other, but in fact their genetic diversity is like the difference between human and invertebrate sea squirt,” said Riley. “We sequenced these diverse genomes to discover and facilitate the next generation of biotechnological workhorse yeasts for producing the fuels and products we use in daily life. We also discovered a genetic code change that, if not understood, will impede the yeasts’ biotechnological use.”
Genetic code reassignment
In P. tannophilus, the team found a change in one of the three-letter codons that represent one of the 20 regularly used amino acids. That change from CUG-Ser to CUG-Ala is only the second observed case of a non-stop codon reassignment (a change from one amino acid to another, rather than from one amino acid to a stop codon) in nuclear genomes. “While we don’t know why and how this happened,” said Grigoriev. “Genes with CUG codons may not produce functional proteins when expressed in an organism with different genetic code as will code for a different amino acid.”

Robert Riley spoke about finding the CUG-Ser to CUG-Ala change in a yeast genome at the 2016 DOE JGI Genomics of Energy & Environment Meeting. Watch his talk at http://bit/ly/JGI2016Riley.
“The CUG-Ala reassignment is important to biotechnology because in order to express novel biotechnologically useful genes from diverse yeasts into workhorses like Saccharomyces, we need to know if the yeasts’ genetic codes are the same,” said Riley. “If they aren’t, expressing the novel genes won’t work because the proteins will be incorrectly translated.”
“With the advent of new genetic tools that can rapidly manipulate an organism’s DNA, publication of these new genomic yeast sequences will open up many new platforms for bioengineering cellulose degrading, lipid producing, acid tolerant yeasts that use a wide range of substrates and produce many different primary and secondary metabolites,” said Jeffries. “The yeast scientific community owes tremendous thanks to the sequencing and annotation team at JGI.”
First author Robert Riley talked about finding a fungus in which the 3-letter codon CUG is interpreted as the amino acid serine rather than the expected lysine during the 2016 DOE JGI Genomics of Energy & Environment Meeting. Watch his talk at http://bit.ly/JGI2016Riley.