Comparative analysis of Aspergillus species provides genus-wide view of fungal diversity
In the world of fungi, Aspergillus is an industrial superstar. Aspergillus niger, for example, has been used for decades to produce citric acid—a compound frequently added to foods and pharmaceuticals —through fermentation at an industrial scale. Other species in this genus play critical roles in biofuel production, and plant and human health. Since the majority of its 350 species have yet to be sequenced and analyzed, researchers are still at the tip of the iceberg when it comes to understanding Aspergillus’ full potential and the spectrum of useful compounds they may generate.
In a study published February 14, 2017 in the journal Genome Biology, an international team including researchers at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, report sequencing the genomes of 10 novel Aspergillus species, more than doubling the number of Aspergillus species sequenced to date. The newly sequenced genomes were compared with the eight other sequenced Aspergillus species. With this first ever genus-wide view, the international consortium found that Aspergillus has a greater genomic and functional diversity than previously understood, broadening the range of potential applications for the fungi considered one of the most important workhorses in the biotechnology.
“Several Aspergillus species have already established status as cell factories for enzymes and metabolites. However, little is known about the diversity in the species at the genomic level and this paper demonstrates how diverse the species of this genus are,” said study lead author Ronald de Vries of the Westerdijk Fungal Biodiversity Institute in the Netherlands. “One can’t assume that an Aspergillus species will have the same physiology as a better studied species of the genus.”
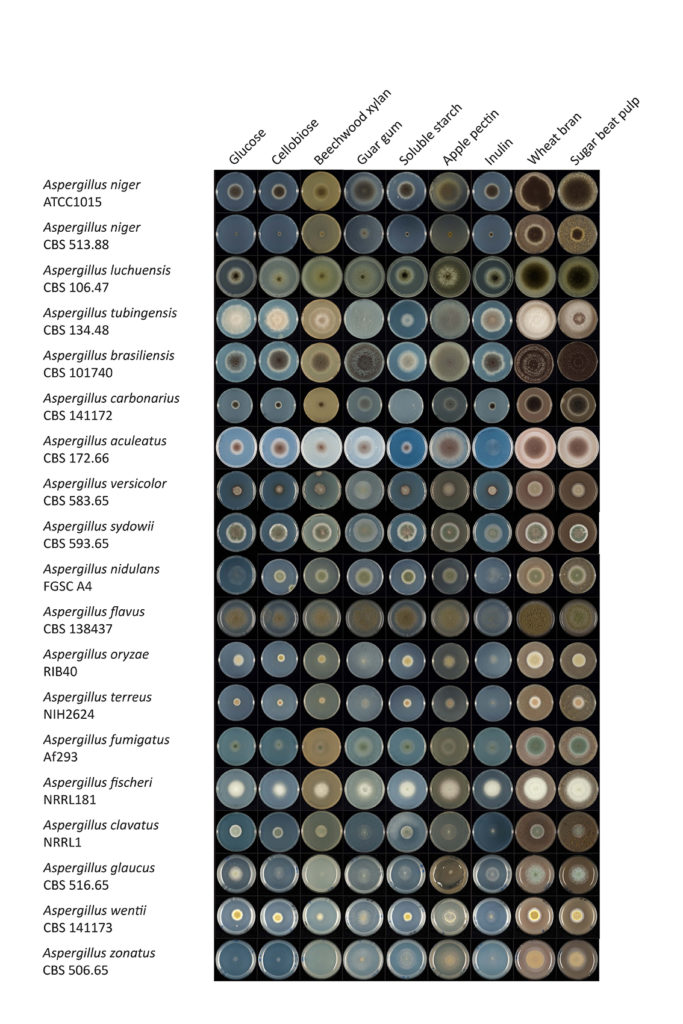
Comparative growth of aspergilli. (Ad Wiebenga & Ronald de Vries, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands)
The study, conducted through the DOE JGI’s Community Science Program, also demonstrates the importance of evaluating biodiversity within a genus to understand how fungi can be greater utilized to solve a variety of problems.
A Catalog of Enzymes for Biotechnological Applications
Sequencing a diverse set of Aspergillus genomes allows researchers to build a more comprehensive catalog of enzymes for biotechnological applications, added DOE JGI Fungal Genomics Program Head Igor Grigoriev, senior author of the paper. Those applications include harnessing Aspergillus to help protect crops and ward off agents that can cause diseases in plants.
Comparing the newly sequenced genomes to those already available, researchers found a huge variety of carbohydrate-active enzymes (CAZymes) among the Aspergillus species, suggesting distinct strategies to break down plant biomass. CAZymes are responsible for breaking down plant cell walls, useful for industrially processing plants the DOE considers candidate bioenergy crops. The sugars that are part of these cell walls can’t be accessed and fermented to make biofuels unless the walls are broken down by agents like CAZymes.
“Each of these 10 genomes encodes for a unique composition of CAZymes—and the wider assortment helps formulate enzyme cocktails better suited for different types of plant biomass to efficiently convert them into biofuels,” Grigoriev said.
In addition to biofuels, CAZymes can also help facilitate the production of paper, textiles, food, feed and pharmaceuticals, according to de Vries.
Pursuing a Deeper Exploration
The comparative analysis between the genomes also enabled researchers to uncover a high diversity of genes that: 1) allow the fungi to produce secondary metabolites, compounds that may be useful for applications such as crop protection; and, 2) enable the fungi to tolerate stress. The knowledge gained in secondary metabolism and stress response will help to provide more insight in the mechanisms underlying these functions.
But despite the new insights gained, de Vries emphasizes that much still remains unknown about the full spectrum of what Aspergillus can do. “There is [still] much to learn and get from a better study [of Aspergillus],” he said. “The potential for applications within the genus has barely been touched.” Grigoriev adds: “Encouraged by results of this study we now pursue a deeper exploration of the Aspergillus genus, sequencing the remaining 300 species, each carrying a unique composition genes, enzymes, and pathways.”
All of the fungal genomes are available on the DOE JGI’s Fungal Genomics portal MycoCosm. Letters of Intent are currently being accepted for the DOE JGI Community Science Program (CSP) and must be submitted online by March 31, 2017. The CSP Annual Call is focused on large-scale sequence-based genomic science projects that address questions of relevance to DOE missions in energy security and sustainability and global biogeochemistry. Click here for more information regarding the CSP call.