WALNUT CREEK, CA—The bane of military quartermasters may soon be a boon to biofuels producers. The genome analysis of a champion biomass-degrading fungus has revealed a surprisingly minimal repertoire of genes that it employs to break down plant cell walls, highlighting opportunities for further improvements in enzymes customized for biofuels production. The results were published online May 4, 2008 in Nature Biotechnology by a team of government, academic, and industry researchers led by the U.S. Department of Energy Joint Genome Institute (DOE JGI) and Los Alamos National Laboratory (LANL).
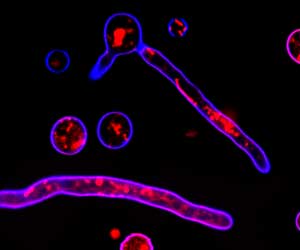
Micrograph of T. reesei hyphae with vesicle membranes stained red and cell wall chitin in blue, courtesy of Mari Valkonen, VTT Finland.
The discovery of Trichoderma reesei, the target of the published analysis, dates back to World War II, when it was identified as the culprit responsible for the deterioration of fatigues and tents in the South Pacific. This progenitor strain has since yielded variants for broad industrial applications and is known today as an abundant source of enzymes, particularly cellulases and hemicellulases, currently being explored to catalyze the deconstruction of plant cell walls as a first step towards the production of biofuels from lignocellulose.
“The information generated from the genome of T. reesei provides us with a roadmap for accelerating research to optimize fungal strains for reducing the current prohibitively high cost of converting lignocellulose to fermentable sugars,” said Eddy Rubin, DOE JGI Director and one of the paper’s senior authors. “Improved industrial enzyme ‘cocktails’ from T. reseei and other fungi will enable more economical conversion of biomass from such feedstocks as the perennial grasses Miscanthus and switchgrass, wood from fast-growing trees like poplar, agricultural crop residues, and municipal waste, into next-generation biofuels. Through these incremental advances, we hope to eventually supplant the gasoline-dependent transportation sector of our economy with a more carbon-neutral strategy.”
For millennia, civilization has long relied on nature’s bounty for shelter and sustenance, with cheap and plentiful supplies of fossil fuels powering the economic engine of the industrial age, leading to the broad diversity of products synthesized from petroleum. With rising concern about dependence on imported oil for transportation, the 21st century is signaling a shift towards “white” or industrial biotechnology—harnessing the metabolic processes of microbes to address energy challenges.
The research team compared the 34-million-nucleotide genome of T. reesei with 13 previously characterized fungi and discovered something counterintuitive. Despite its reputation as an avid plant polysaccharide degrader, T. reesei, was found to have the smallest inventory of genes powering its robust degradation machinery.
“We were aware of T. reesei’s reputation as a producer of massive quantities of degrading enzymes; however, we were surprised by how few enzyme types it produces, which suggested to us that its protein secretion system is exceptionally efficient,” said Diego Martinez, the study’s lead author and researcher supported by DOE JGI at LANL, and at the University of New Mexico. Subsequently, he and his colleagues turned their attention to the complexities of T. reesei’s secretory pathway components, which they had a hunch played an important role in the organism’s success.
“While little appears to have changed in the secretion machinery since divergence with a common ancestor with yeast,” said Martinez, “there are some intriguing differences in the way T. reesei processes some protein bonds important for cellulase production.”
In their comparative analysis of T. reesei with other fungi, the team observed clustering of carbohydrate-active enzyme genes, which suggested a specific biological role: polysaccharide degradation. “While plant tissues are not likely the main source of nutrients for T. reesei, upon detection of cellulose and hemicellulose it seems that the organization of these degrading genes may be the key to a rapid response,” said Martinez.
“The sequencing of the Trichoderma reesei genome is a major step towards using renewable feedstocks for the production of fuels and chemicals,” said Joel Cherry, director of research activities in second-generation biofuels for Novozymes, one of the collaborating institutions on the study. “This soft rot fungus serves as the world’s most prodigious producer of cellulases and is already a dominant source of a wide variety of cellulase products for the textile industry worldwide. It is also the organism of choice for producing enzymes for the breakdown of cellulosic biomass to fermentable sugars, which can then be biologically converted to fuels and chemical building blocks. The information contained in its genome will allow us both to better understand how this organism degrades cellulose so efficiently and to understand how it produces the required enzymes so prodigiously. Using this information, it may be possible to improve both of these properties, decreasing the cost of converting cellulosic biomass to fuels and chemicals.”
Other authors on the study include, from JGI-LANL (at Los Alamos National Laboratory), Thomas Brettin, David Bruce, Chris Detter, Cheryl Kuske, Olga Chertkov, Melissa Jackson, Cliff Han, Monica Misra, Nina Thayer, Ravi Barbote, and Gary Xie; from the JGI-PGF (Production Genomics Facility in Walnut Creek, CA), Jarrod Chapman, Igor Grigoriev, Isaac Ho, Susan Lucas, Nicolas Putnam, Paul Richardson, Daniel Rokhsar, Asaf Salamov and Astrid Terry; and Pacific Northwest National Laboratory’s Scott Baker and Jon Magnuson. Other collaborating institutions include the USDA Forest Products Lab, Oregon State University, University of New Mexico, TU-Vienna, Catholic University of Chile, VTT of Finland, and Universités d’Aix-Marseille I & II.
The U.S. Department of Energy Joint Genome Institute, supported by the DOE Office of Science, unites the expertise of five national laboratories — Lawrence Berkeley, Lawrence Livermore, Los Alamos, Oak Ridge, and Pacific Northwest — along with the Stanford Human Genome Center to advance genomics in support of the DOE missions related to clean energy generation and environmental characterization and cleanup. DOE JGI’s Walnut Creek, CA, Production Genomics Facility provides integrated high-throughput sequencing and computational analysis that enable systems-based scientific approaches to these challenges.