 WALNUT CREEK, CA — The streamlined genome of Ciona intestinalis, a common sea squirt closely related to vertebrates on the evolutionary tree, is providing new clues about the origins of key vertebrate systems and structures including the human hormone, nervous and immune systems.
WALNUT CREEK, CA — The streamlined genome of Ciona intestinalis, a common sea squirt closely related to vertebrates on the evolutionary tree, is providing new clues about the origins of key vertebrate systems and structures including the human hormone, nervous and immune systems.
In an article for the December 13, 2002 edition of the journal Science, an international consortium of researchers reports on the draft sequencing, assembly, and analysis of the genome of C. intestinalis. The consortium is led by the U.S. Department of Energy’s Joint Genome Institute (JGI); the Department of Molecular and Cellular Biology and Center for Integrative Genomics at the University of California, Berkeley; the Department of Zoology at Kyoto University, Japan; and Japan’s National Institute of Genetics in Mishima. It includes nearly two dozen other research institutions.
By comparing Ciona‘s genome with those of the human and other animals, the researchers were able to glean new insights into the evolutionary origins of the human brain, spine, heart, eye, thyroid gland, and nervous and immune systems, as well as a better understanding of chordate and vertebrate development in general.
“We have known for years that despite their humble appearance, sea squirts, or ascidians, have a number of physical characteristics that are similar to vertebrates,” said JGI Director Eddy Rubin. “Through a comparison of their genomes, we can now understand how these relationships are reflected at the molecular level, and how these similar systems and gene sets evolved in different ways over 500 million years from a common ancestor.”
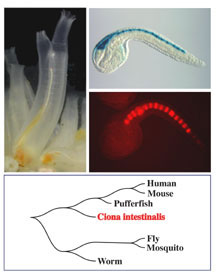
The sea squirt is a urochordate, or “tunicate,” found in shallow ocean waters around the world. The barrel-shaped adult squirt attaches to rocks, piers, boats and the sea bottom, and feeds by siphoning seawater through its body and using a basket-like internal filter to capture plankton and oxygen.
It is as juveniles, however, that ascidians reveal their kinship to humans. One day after an egg is fertilized, it develops into a small tadpole comprised of only about 2,500 cells. The tadpole soon finds a home, settles down, and metamorphoses into its immobile adult form — but while still in its swimming stage the tadpole has a stiffened rod running the length of its tail called a notochord, the forerunner of our backbone, as well as a primitive nervous system.
Ciona‘s close relationship to vertebrates, along with its compact genome — about 160 million base pairs, one-twentieth the size of the human — and small cell complement make it an ideal model organism for studying chordate development and DNA regulatory networks. The JGI’s comparative genomic studies of multiple organisms, which are funded by the Office of Biological and Environmental Research in DOE’s Office of Science as part of its Human Genome Program, are helping to determine the functions and regulation of the genes and other DNA regions in the human genome that play a crucial role in human health.
The Ciona sequence analysis revealed that its genome contains about 16,000 genes, about 80 per cent of which are also found in humans and other vertebrates. The total number of Ciona genes, however, is only about half the number in vertebrates. Daniel Rokhsar, head of the JGI’s computational genomics department, said one reason for this “slimmed-down” DNA sequence is the fact that Ciona has single copies of a large number of genes that are present in multiple copies in vertebrates.
“One of two things can happen to these redundant copies,” Rokhsar said. “Either they mutate away and disappear, or they evolve to perform other functions. By looking at Ciona‘s genome, we can see what innovations occurred in the human lineage that enabled us to advance in complexity.”
For example, the Ciona analysis revealed a number of vertebrate-like genes, including complement, lectin, and Toll-like receptor genes, that researchers said may play a role in Ciona‘s primitive “innate” immune system. The scientists found no evidence, however, of the hundreds of key genes, such as immunoglobulin or T cell receptors, involved in the adaptive immune systems found in higher vertebrates.
Ciona is also missing a number of the important Hox genes that determine vertebrate body structure. In some cases only single Hox genes are present, while some appear to be entirely absent. On the other hand, Ciona has light-sensing genes very similar to those of vertebrates, as well as genes involved in the formation of the heart and of cells that accumulate iodine, much like the human thyroid gland. Ciona, however, uses a protein called hemocyanin — rather than the hemoglobin used by most other animals — to transport oxygen through its bloodstream, and it lacks the genes to produce steroids and histamines.
The researchers were surprised to find that in some ways, Ciona has more in common with bacteria, fungi and plants than with vertebrates: the genes it uses to produce the fibrous tunic that supports its feeding siphons are similar to the genes involved in metabolizing cellulose. “Because cellulose is typically produced only by plants and bacteria, its presence in ascidians is a curious lineage-specific evolutionary innovation,” the researchers said.
“Just like vertebrates, modern ascidians have been evolving in their own way from our common origins,” said Rokhsar. “The Ciona genome reflects the ways they have mobilized many of the same genetic raw materials that we both inherited to serve their own specialized needs.”
Mike Levine, professor of genetics and development at UC Berkeley, noted that the Ciona sequence analysis is “entirely consistent” with Darwin’s 1871 suggestion in The Descent of Man that ascidians and vertebrates diverged from a common ancestor, with, as Darwin put it, “…one branch retrograding in development and producing the present class of Ascidians, the other rising to the crown and summit of the animal kingdom by giving birth to the Vertebrata.”
Ciona was sequenced using the whole-genome shotgun method, which has also been used to sequence the human, pufferfish, mouse, and rat genomes. Earlier this year the JGI convened a Ciona “annotation jamboree” in Walnut Creek, bringing together an international group of experts from the United States, Japan, Italy, France, Scotland, Canada, and Australia to analyze the sea squirt genome and begin disentangling its evolutionary relationship with other animal genomes.
“Bringing the community together was essential,” Rokhsar said. “Only by a collaborative effort could we hope to get a coherent sense of the relationship of the Ciona to other organisms, which in turn illuminates the mechanisms of evolution.”
In addition to analyzing the Ciona‘s genome, JGI researchers are also conducting experiments to help identify the function of non-coding sequences of DNA lying between the genes, which regulate gene expression and determine when and where in the organism a given gene is turned on and off. Understanding the complex orchestration of gene networks is crucial to much of contemporary biomedical research. Results of those studies are expected to be published in early 2003. Meanwhile, Ciona intestinalis sequence information and additional information on the sea squirt can be accessed on JGI’s website at http://genome.jgi.doe.gov/ciona.
The JGI, established in 1997, is one of the largest and most productive publicly funded human genome sequencing institutes in the world. The JGI was founded by three DOE national laboratories managed by the University of California: Lawrence Berkeley National Laboratory and Lawrence Livermore National Laboratory in California and Los Alamos National Laboratory in New Mexico. In addition to the Ciona project, the JGI has whole genome sequencing programs that include vertebrates, fungi, plants, and bacteria and other single-celled microbes. Funding for the JGI is predominantly from the Office of Biological and Environmental Research in DOE’s Office of Science, with additional funding from NIH, NSF, USDA and NASA. Additional information and progress reports on JGI projects, including daily updates of sequence information and assembly statistics, are available at www.jgi.doe.gov.
The National Institute of Genetics (NIG) is located in the city of Mishima near Fuji Hakone National Park. It was established in 1949 as the central institute for studies on various aspects of genetics. In 1984, NIG was reorganized as an inter-university research institute to promote collaborative studies. NIG serves as a stock center for various genetic resources and for the DNA Data Bank of Japan, and provides excellent research activities for inter-university collaborations. In 1988, the Graduate University for Advanced Studies was founded, and NIG now undertakes graduate education at the Department of Genetics. NIG has been exploiting the evolving nature of genetics to extend the frontiers of the life sciences.