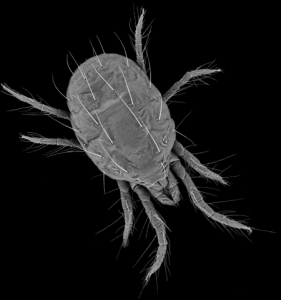
WALNUT CREEK/BERKELEY, Calif.—For a pest that isn’t quite the size of a comma on a keyboard, the two-spotted spider mite can do a disproportionate amount of damage. These web-spinners extract the nutrients they need from leaves of more than a thousand different plant species, including bioenergy feedstocks and food staples. The cost of chemically controlling spider mites to counteract reduced harvest yields hovers around $1 billion annually, reflecting their significant economic impact.
With a 90-million nucleotide genome, the smallest of those that belong to the group of animals with external skeletons or arthropods, the two-spotted spider mite was selected for sequencing in 2007 by the U.S. Department of Energy (DOE) Joint Genome Institute (JGI). “Many aspects of the biology of the spider mite seem to facilitate rapid evolution of pesticide resistance,” said DOE JGI collaborator Yves Van de Peer of the Flemish Institute for Biotechnology (VIB) and Ghent University, Belgium. “Control of these mites has become increasingly difficult and the genetic basis of such resistance remains poorly understood.”
Van de Peer and others in the research community are now employing the publicly available genomic data from the spider mite to advance the development of novel pest-control strategies that could serve as an alternative to chemical pesticides and reduce environmental pollution. In the November 24 edition of Nature, he and an international team of researchers from more than 30 institutions reported on how the spider mite is shedding light on questions such as the pest’s ability to rapidly develop resistance to pesticides, how they can serve more broadly as a model for pest-plant interactions and how they are likely to respond in a changing environment.
“The analysis revealed mechanisms underlying such diverse traits as pest-plant interactions inspiring novel crop plant protection strategies, and the evolutionary innovation of silk production, presenting opportunities for new nanoscale biomaterial development,” said the publication’s first author Miodrag Grbic of the University of Western Ontario, Canada, and the Instituto de Ciencias de la Vid y el Vino, Logroño, Spain. Grbic was also the project lead in proposing that the spider mite, Tetranychus urticae, be sequenced under the DOE JGI’s Community Sequencing Program (CSP). Additional support came from Genome Canada and the Ontario Genomics Institute.
“From a pest management perspective, our colleagues are applying these data as the basis for predicting the effects of climate change on the biology, distribution, and abundance of T. urticae, and as a model system, stimulate advances in similar research for other arthropods,” Van de Peer said. Damage of crops by pests represents one of highest energy losses in agricultural production. A tremendous amount of energy is invested in soil tilling, seed distribution, and fertilizer, pesticide, and herbicide application—not counting the energy consumed in the production of those fertilizers and pesticides. Spider mite infestation typically occurs at the late stages of crop development, causing wilt and subsequent degradation that leads to losses of crop and all the associated agricultural inputs.
Of the estimated two million species of mites, less than five percent have been described in any detail. Of this fraction, the spider mite T. urticae is the first of the chelicerate group, which includes spiders, scorpions, and horseshoe crabs, to have its genome completed. Among the arthropods, the tiny pest joins the water flea Daphnia pulex, the first crustacean to have its genome sequenced and published earlier this year by DOE JGI and its collaborators, in the expanding portfolio of biologically-important model systems.
Another discovery associated with the published work was the characterization of genes transferred between species. “It’s exciting to identify microbial and fungal genes that have been incorporated into the spider mite genome,” said author Jeremy Schmutz, leader of the DOE JGI Plant Program (and faculty investigator at HudsonAlpha Institute for Biotechnology). “It adds evidence supporting the theory of lateral gene transfer as mechanism for plant pathogens to specialize on plants and increase the ability of their population’s impact on our food sources.” Schmutz goes on to say that part of a larger strategy devised to supplant the use of fossil fuels includes a push to plant out large quantities of cellulosic crops for biofuels, which requires a better understand of the plant interactions with major groups of pathogens. “Most of the genomic work has been done with molds and fungal pathogens, but insects are a major issue for biofuel crops.”
The availability of the mite’s genome is also fueling research in the biomedical arena, the authors note, including developmental studies characterizing the infectious mechanisms in related organisms, such as ticks that are vectors for Lyme disease and hemorrhagic fever and other mites that trigger allergic responses.
“Like the fruit fly Drosophila and the worm C. elegans, these organisms are rallying points around which pioneering research takes shape,” said Jim Bristow, DOE JGI Deputy Director of Science Programs who also oversees the CSP. “It wasn’t too long ago when Daphnia had only a handful of dedicated researchers. Now, with the water flea genome available, and its recognition as a keystone species in freshwater ecosystems on the rise, Daphnia research groups number in the hundreds worldwide. This is among our goals in making this information available, to catalyze such energy- and environmentally-relevant fundamental research.”
Supported by the Office of Biological and Environmental Research in the DOE Office of Science, the DOE JGI’s Community Sequencing Program enables scientists from universities and national laboratories around the world to probe the hidden world of microbes and plants for innovative solutions to the nation’s major challenges in energy, climate, and environment. Follow the DOE JGI on Twitter and Facebook.
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.