Demonstrating the microfluidic-based, mini-metagenomics approach on Yellowstone hot springs samples.
The Science
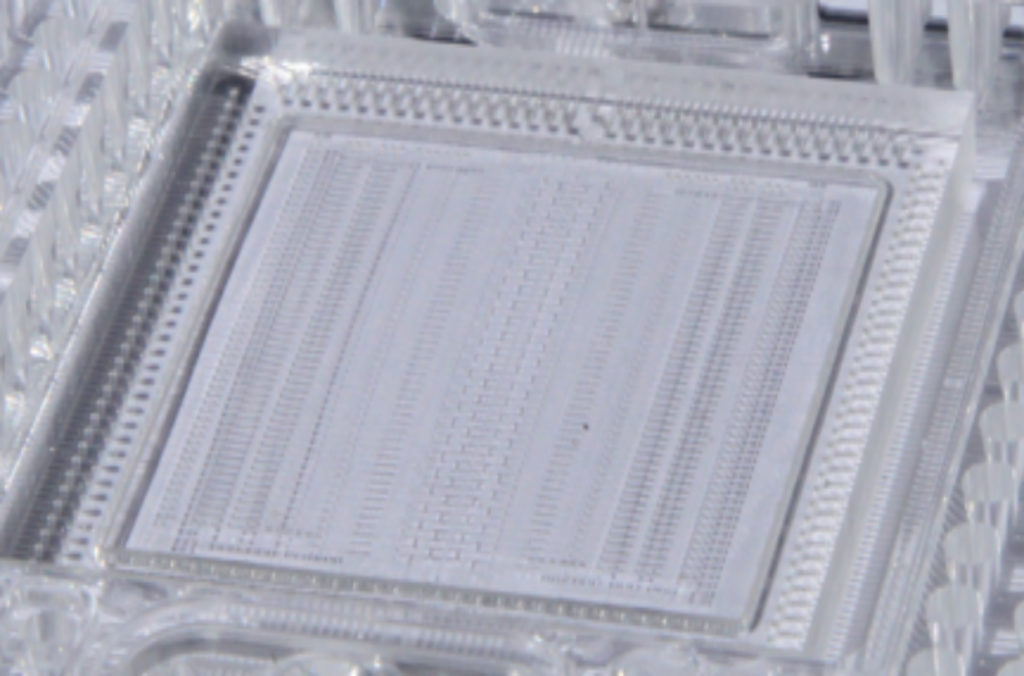
Integrated microfluidic circuit used to perform mini-metagenomic microbial cell partitioning and genomic DNA amplification. (Brian Yu)
Stanford researchers extracted 29 novel microbial genomes from Yellowstone hot spring samples while still preserving single-cell resolution to enable accurate analysis of genome function and abundance. The work was enabled by the Emerging Technologies Opportunity Program (ETOP) of the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility. Letters of Intent are currently being accepted for the FY18 ETOP and are due by August 30, 2017 – click here to learn more.
The Impact
New technologies that can be harnessed to illuminate so-called microbial “dark matter,” the majority of the planet’s microbial diversity that remains uncultivated, have many benefits. The genes and metabolic functions of these yet undiscovered microbes enable them to live in a wide range of environments, and could have potential applications in fields ranging from bioenergy to biotechnology to environmental research.
Summary

Samples used to demonstrate the efficacy of the new technology were taken from hot springs at Yellowstone National Park. (Image by Paul Blainey, Christina Mork and Geoffrey Schiebinger)
There are more than 50,000 microbial genome sequences in the DOE JGI’s Integrated Microbial Genomes (IMG) publicly accessible database, and many of them have been uncovered through the use of metagenomic sequencing and single-cell genomics. Despite their utility, these techniques have limitations: single-cell genome amplifications are time-consuming, often incomplete, and shotgun metagenomics sequencing generally works best if the environmental sample is not too complex. In eLife, a team of Stanford University researchers reported the development of a microfluidics-based, mini-metagenomics approach to mitigate these challenges. The technique starts with reducing the environmental sample’s complexity by separating it using microfluidics into 96 subsamples of 5-10 cells. Then the genomes in the few cells in each subsample are amplified and libraries are created for sequencing of these mini-metagenomes. The smaller subsamples can be held to single-cell resolution for statistical analyses. Co-occurrence patterns from many subsamples can also be used to perform sequence independent genome binning. The technology was developed through resources provided by the DOE JGI’s ETOP, which was launched in 2013. The aim of ETOP is to use these new technologies to tackle energy and environment applications, adding value to the high throughput sequencing and analysis currently being done for DOE JGI users. The team validated the technique using a synthetic microbial community, and then applied it to samples from the Bijah and Mound hot springs at Yellowstone National Park. Among their findings was that the microbes at Mound Spring had higher potential to produce methane than the microbes from Bijah Spring. They also identified a microbial genome from Bijah Spring that could reduce nitrite to nitrogen. Applying this new technology to additional sample sites will add to the range of hitherto uncharacterized microbial capabilities with potential DOE mission applicability.
BER Contact
Daniel Drell, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contacts
Tanja Woyke
Microbial Program Head
DOE Joint Genome Institute
[email protected]
Stephen Quake
Department of Bioengineering
Stanford University
[email protected]
Funding
Work was conducted by the U.S. Department of Energy (DOE) Joint Genome Institute, a DOE Office of Science user facility (contract number DE-AC02-05CH11231). This work was also supported by the John Templeton Foundation, Stanford University, National Science Foundation, and the Burroughs Wellcome Fund.
Publication
- F.B. Yu, P. C. Blainey, F. Schulz, T. Woyke, M. A. Horowitz, and S. R. Quake. “Microfluidic-based mini-metagenomics enables discovery of novel microbial lineages from complex environmental samples.” eLife. 2017 July 5. DOI: 10.7554/eLife.26580
Related Links
- DOE JGI Emerging Technologies Opportunity Program
- FY2018 ETOP Call for Letters of Intent
- Phylogenetic Diversity, a mission of the DOE JGI Microbial Program
- DOE JGI News Release: “Boldly Illuminating Biology’s ‘Dark Matter’”
- DOE JGI Science Highlight: “Confirming Microbial Lineages Through Cultivation-Independent Means”
- DOE JGI Science Highlight: “Elucidating Extremophilic ‘Microbial Dark Matter’”
- DOE JGI Science Highlight: “Solving Microbial ‘Dark Matter’ With Single-Cell Genomics”
- “Why Mine Microbial Dark Matter?” video
- Video of Slava Epstein of Northeastern University on the importance of studying microbial dark matter
- Video of Steve Quake at the 2014 DOE JGI Genomics of Energy & Environment Meeting