Enediynes, enabling technologies, and leveraging a large microbial strain collection for natural product discovery.
This is our conversation with Professor Ben Shen, from The Scripps Research Institute (TSRI) in Jupiter, Florida. Ben and Alison and I talk about enediynes and their use in medicine, how Ben got fascinated with natural products by working on terpene chemistry, TSRI’s acquisition of the Pfizer strain collection, and our collaborations to sequence that collection, mine genomes, and develop new technology to access natural products.
Show Notes

As promised in the intro to Episode 7, here is some additional info about “enediynes”, one of the most toxic classes of natural products known!
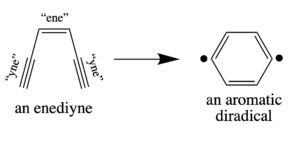
The name comes from their core chemical structure: one “ene” – that’s two carbons attached by a double bond – and to “ynes” – two more pairs of carbons attached by triple bonds, each one attached to one end of the double bonded carbons. This is hard to say coherently, and harder to type, so here’s a drawing.
On its own, this is a really unstable configuration of carbons, and, instead, these six carbon really want to rearrange into a six-membered aromatic ring. When they do that, they end up creating a “di-radical” which can wreak reactive havoc on almost any other molecule. However, in the natural product versions of enediyne molecules, there’s a whole bunch of other atoms that can either stabilize the enediyne, and/or provide the chemistry needed to direct this unstable warhead to its target.
As we discuss in the podcast, enediynes are often used as the “payload” on an antibody-drug conjugate, or “ADC”. The enediynes that we find in nature weren’t evolved for the purpose of killing human cancer. But, some are very toxic and can be used that way – the main side problem being that they also kill regular cells too! So, scientists figured out that if you take an antibody that can recognize and bind to cancer cells, and then you attach a toxic molecule – like a natural product enediyne – to it, then the antibody will bring the toxic molecule straight to the cancer cells and the toxin can kill it. Teamwork!
If you’re somewhat knowledgeable in natural products or biosynthetic chemistry and want to dive deep on the topic, or just want to take a look at several natural product enediyne structures, Ben’s 2015 paper on genome mining for enediynes is one of my favorites.
Transcript
DAN: You’re listening to the US Department of Energy Joint Genome Institute’s “Natural Prodcast,” a podcast about the science and scientists of secondary metabolism.
DAN: Hello, and welcome back for Episode 7 of the Natural Prodcast. It’s been about a month now since we dropped our first six episodes, and now after gathering some feedback and making a few technical tweaks, I think we’ll be on a pretty regular schedule for a while. Moving forward, I’m hoping to get a new one out every two weeks. This time, we’ve got a great interview with Ben Shen, from the Scripps Research Institute in Jupiter Florida. This interview is another one we recorded at the Society for Industrial Microbiology Natural Products conference that was held last January. Ben was the organizing chair of that conference, and he and the conveners put on a great program. This is one of my favorite interviews so far – you’re going to hear how passionate and enthusiastic Ben is about our field, and I think his energy is infectious.
DAN: I’ll say up front that JGI recently started collaborating with Ben and his group to work on sequencing, synthesis and genome mining technology to help access complicated natural product gene clusters, and we’re all really excited about that, and I think that comes across in the interview. If you listened to the Primer episodes, then you heard me talk about JGI’s Earth Secondary Metabolome initiative, and our collaboration with Ben is an important part of that.
I’m also going to mention that Ben talks fast – it’s part of his energy – and he has an accent, and, so, I don’t really have any trouble understanding him, but I know from the download stats that we actually have a lot of listeners downloading this from countries all over the world – as many from outside the US as inside – and so, especially if English is not your first language, if there’s ever anything you didn’t catch in the audio, head over to naturalprodcast.com, where you’ll find transcripts and show notes for every episode we put out. I actually put a lot of time into preparing those transcripts, so I hope somebody’s going to find them useful!
DAN: Among many other things, Ben is maybe best known as an expert in enediynes – that’s a class of natural product with a really cool chemical structure. We try to explain it, but it’s chemistry, so I don’t think audio really conveys what’s going on there. So, check out the show notes, and I’ll put up some structures and link to some resources you can use to get caught up, if you’re not already familiar with them.
DAN: So, here we go. Natural Prodcast episode 7, our talk with Ben Shen.
DAN: Alison, we’re not in Berkeley. I’m not in Paris.
ALISON: Oh my god. Where are we? I know, I know. I know.
DAN: You know where you are, I hope?
ALISON: Oh, yeah, yeah..
DAN: Yeah, we’re in San Diego! We’re at the SIMB natural products conference. And today we’re going to talk to Ben Shen!
ALISON: Awesome. Yeah, I’ve heard a lot of good things about Ben. Nice to meet you, Ben.
BEN: Hey, yeah! Great, I’m very happy to talk to you. And so this is the third international meeting on natural products, but as a new, I call it “incarnation”, for this series of meetings. It’s because – mainly it’s due to the emerging technology on DNA sequencing and synthetic biology. But also the metagenomics and gut microbiome, as I think that it’s like a perfect storm in terms of fundamental knowledge and enabling technology, but interesting biology and a potential application of translation in medicine. So it’s a really great time to convene, you know, people all over the word who have a common interest, united by natural products,
DAN: You guys have put together a fantastic program. So I’m looking forward to the next couple of days. Yeah, yeah. So you are at the Scripps Research Institute. We’ve known each other for a while, you are definitely an expert in natural products. And I’ve always known you as “the enediyne guy”. And so enediynes are this really cool class of natural product compounds, Alison. They are – if you can imagine – two carbons in a triple bond attached to two carbons in a double bond attached to two carbons in a triple bond. And then you know, the rest of a natural product molecule around that. That’s sort of a semi-stable configuration of electrons that becomes really reactive. And so, different types of enediyne molecules can bind to different places, but then the ring opening can cause all kinds of crazy havoc depending on what the drug wants to do.
ALISON: Okay, so it’s for drug applications. Is it any particular kinds of drugs? Like cancer-fighting, or…?
BEN: Yeah. So, if I want to put it into perspective, right, so the enediyne structure – the enediyne compound that people know, as a family, it’s more than 7 decades. So, this family of substances is always associated with toxicity, basically, killing cells.
ALISON: Right, because they’re so reactive.
BEN: …going back to the 1950s. But the structure of which was not known until the middle ’80s. And then the first time we saw the structure that Dan just described to you. So the enediyne referred to two carbon with a double bond. And “di” means two and it means two carbons with triple bonds. So it’s a double bond flanked by two triple bonds. So this is called enediyne. And what is unique about enediynes, is really another example. This sort of carbon arrangement, which we know as a structure of a natural product, was not known to the chemists. So that’s some of the beauty of natural products – it inspires in the fundamental aspects of it. If there were not natural products to teach us about the enediyne structure, we would never know that carbon can connect in such a way to give us the structure. So that’s the fundamental aspects. And the second, from a biological potential application perspective, is because this compound when they were first structurally resolved in the 1980s, they were known as the most toxic molecules, and that human beings know, today, for small molecules. So they’re in the picomolar range.
ALISON: Picomole… is [10 to the] minus twelve, right?
BEN: Yeah, yeah. Well, we are at [10 to the] minus 12 [Molar concentration]- is sufficient to kill cells. So it’s really remarkable in terms of a novel mechanism, how a small molecule can kill. So that immediately inspired people to make use of this molecule for biomedical application. So the Yeah,
ALISON: But how? I mean, it’s so toxic…
BEN: Yeah, yeah. So this molecule is so toxic. So people have tried this for a long, long time. So if you think about it, because the compound is so toxic, and the mechanism, which is breaking DNA, so there’s no selectivity. So, people have been thinking about the chance to deliver this molecule – very toxic molecule – to specific disease cells. So, the very first compound was introduced in the early 90s and by using a polymer as a delivery mechanism. So, the polymer structure has specific preferences to cancer cell lines. So, that was introduced in Japan as a very first example. So the enediyne was introduced clinically. And then the major application has come from Pfizer’s introduction of one of the enediyne compounds by coupling it with antibodies.
DAN (post edit): Hey, Dan here with a quick interjection. Ben asked me to correct what he just said here. It was actually the drug company Wyeth that introduced enediynes to the market in 2001 with a drug called Mylotag. Wyeth was later acquired by Pfizer in 2009. So, a simple mistake, but I’m happy to be able to correct the record.
BEN: So, antibody is one of the macromolecules that can specifically recognize a specific cell type. So by using the antibody as the recognition motif, coupled with the enediynes, the disease cell killing ability, and we have this, called antibody drug conjugates, as a new type of medicine that will open up a new year for targeted anti cancer drug discovery.
DAN: Yeah, great. Are any of these antibody-drug conjugate enedynes used in the clinic now?
BEN: Yeah, so the very first one, first that we call antibody drug conjugates, abbreviated as “ADC”, was approved in the year 2000, and utilize the enediyne as the payload. We call it payload – payload means the drug, and the antibody used as a vehicle to deliver the drug. And up to now there are six ADCs that have been FDA approved over the past two decades, and, remarkably, of the six, two of them utilize the enediyne as a payload molecule. The other four also utilize two different natural products. So a natural product really rocks if you’re thinking about ADCs!
ALISON: ADCs stand for…?
BEN: Antibody Drug Conjugates.
ALISON: Okay, gotcha.
BEN: Yeah. To “conjugate” means to link antibody and drug together.
ALISON: Right. Okay. Okay. Thank you.
BEN: So in the area of ADC, right, if you want to be thinking about natural products… I think two things are very important. Well, two things probably differentiate natural products from many other small molecules, is that natural products always teach us in the principles of biology.
DAN: That’s right.
BEN: You know, to say, man-made compounds. How can you design a drug, right? You design drugs based on certain knowledge, without which, you know… If you asked me, Ben, you discovered -if can make something you have to have some fundamentals. What is beautiful for natural products is nature! Nature does this sort of – if you think of nature as a scientist, nature is doing science, way longer than we as a civilization, supposedly.
DAN: By a few billion years, yeah!
BEN: Yeah by a few billion years. So, there are so many things that we can learn from nature. So, this is from natural product – they also often surprise us by giving us insights, inspires us to discover the fundamentals and then apply the findings and to translate our fundamental findings into specific applications for our human benefits.
DAN: Now, you and I are working together on a project at the JGI to try to find new enediyne clusters and to figure out better ways to produce them. Do you want to talk a little bit about that project and how that got started?
BEN: Yeah, so I want to put this specific project that we are working on into perspective in the field in general. So what has happened in the past two decades is that today, one thing is the ability of genome sequencing. So, in the past the natural products have been discovered simply by isolating the natural product. So the natural product can be in every place, but you will not know they existed, up until you isolate the compound. But today, by genome sequencing, you can predict where in particular areas, or particularly as by ecological niches, the natural product could be there. So this has really changed the paradigm of how to discover natural products. So, why I’m very excited to be working with Dan, you, with the JGI, is that – so JGI can do two things very well. One is to do DNA sequencing. So upon sequencing many genomes, now we don’t have to, as in the past, go randomly looking in places to isolate. And we can now specifically look in places [that we’re] only aware of upon DNA sequencing, we know potentially can make a compound.
DAN: That’s right.
BEN: And then once you know that the DNA is all saying which biological niches from which potentially natural product could be biosynthesized, then you need to develop enabling technologies. ie how to translate the ATGC sequence into discrete small molecules. So, Dan, I’m very excited, and we are using the enediyne for its structural novelty, but also for its potential application in human medicine. And so, we are developing enabling technology, not only to facilitate the discovery of enediyne, for which, by genome sequencing, we know they are more proliferative than the enediyne we have isolated to date by the traditional methods. But more importantly, we hope through enediyne that we will be able to develop much broader applicable technologies that can be applied to help the discovery of other classes of natural product.
DAN: That’s right. Yeah. Yeah, once you learn the technology to, you know, work with one given particular class you might be interested in, you can usually apply that to just about anything. Because, basically we’re working with DNA. Yeah. And in DNA is all that the same molecule, just different sequences
BEN: The other thing I think about JGI is, really, it’s probably the best place to do it. You have the sequencing capacity, you have the bioinformatics, but also you have all the emerging technology under one roof. So that really facilitates. The metabolomics, another technology that to, monitor if all the, say, emerging technology we apply to translate the DNA into the small molecule – if these methods are very effective, that is, if the natural product has been produced. So I think that I’m excited for everything that seems like, you know, falling into the right place.
DAN: Yeah, at the end of the day, you got to get a molecule in a bottle and so that’s…
BEN: In sufficient quantity!
DAN: Sufficient quantity! Yeah, just like we talked about with taxol. [See Episode 1!]
ALISON: So you said, you know, it’s valuable to have all of these technologies and expertise under the same roof. I wondered if you’ve had, like, a negative experience somewhere else, or you know, you’ve run into challenges that demonstrate why this is so important.
BEN: Yeah. So I wouldn’t call it a negative experience. It’s a couple of things for the current research. You have to… I think it’s important. One is many of the technologies – it’s no longer a silo. It really requires interaction from many different disciplines. And so this is where a traditional, like, silo, or compartmentated research, will not work really effectively. The second is that I like the idea with JGI, and national labs, is that, I think any invention will be useless unless you make the invention available to broad user groups. So, I view the project that we are working on together, the mandate is whatever we discover we can make it available to broad audiences. So, not only I say we were able to make use of it, but also the community can benefit from the technology. But even better, the more people who will use your technology and then they can help you to fine tune your technology and probably improve it, but also, because of the broad application, we probably can see it pushing the envelope and improve the technology, but also the scope of the technology, in that aspect.
DAN: So I wanted to ask you also about – because you were in the news recently – TSRI has acquired the Pfizer strain collection. And so that’s a big collection of natural product-producing organisms that Pfizer, you know, mined for years to try to find new active compounds. And we talked about how natural product based or organism extracts are not being used very much anymore in industry. So, what’s the value of that collection to academia?
BEN: I’m so glad, Dan, you’re asking this question. And I am so excited and so passionate about this particular strain collection.
DAN: I do know the answer!
BEN: Yes. Yes. I know you knew the answer, but I want to still give you – just regurgitate why I’m very excited. I think, among many, many major discoveries since the genomic revolution, to me, as a natural part chemist for microbial natural product, is that I love microbial natural products is because one of the traditional limitation for natural products – say if we originated, say, taxol, for example. If you say isolate compound from the trees – if you’re cutting the trees, it takes years to regrow the trees back. And the beauty for microbial natural products is the microorganism, and you can do fermentation, which we know how to do since ancient times!
We know how to ferment beers or wines or rice liquors etc. So, by sequencing the genome of microorganisms, the very first one for the natural product producing organism, the Actinomyces, was completed in early 2000s. One of the major discovery from the genome sequencing of this Actino strains, instead of knowing some of the organism can produce one or two compounds, by looking at the genome – And by looking at the genome, the genome predicts they often can produce around 30 – more than 30 compounds. So, this was a complete surprise. So total, and different people have different numbers, but in general, an authoritative number for total number of natural products of microbial origin is around 30,000 from fungal, and a 40,000 from bacteria. Okay? And among the 40,000 from bacteria, there’s around 20,000 from the Actinomycetes. So, now, from the 20 thousand from Actinomycetes, and among the 70,000 microbial natural products total, about 500 drugs have been out of these natural products. So this gives you approximately a 6% successful drug discovery rate.
And this is 100 times better, on average, in comparison to the drug discovery from man-made campounds. So if you are interested in drug discovery, you would like to follow the winners. So natural products is clearly a winner. But the problem is not lack of effort. We just don’t know how to discover.
So this goes back to the Pfizer collection. So the Pfizer collection – from this collection in total contains approximately 100,000 bacterial strains, and a hundred thousand fungal strains. And if you believe the prediction that each strain will produce – has the potential to produce – 30 compounds, I’m talking about 3 million natural products of bacterial origin and 3 million natural products of fungal origin. Now, if I put this number in reference to the non-natural product, there’s only 30,000 natural products of fungal origin known to the entire civilization. Suddenly I have hundreds of strains that has a potential to produce 100-fold more of all the natural products. If Dan and I were able to develop the enabling technology as we are doing for the enediynes, we can enrich the chemical space of natural products in almost a hundred-fold. That is going to change the world! And that’s why I’m so excited in terms of the strains we are getting.
The other thing is really remarkable. So, as a history, right, if I am using probably a most simple-minded sort of analogy, I would consider Alexander Fleming – the discovery of penicillin from the fungal species – probably told us that fungi could be very productive sources for, say, biological active compound discovery. For this Fleming won the Nobel Prize in the 40s. Then Samuel Waxman taught us the isolation of streptomycin from Actinomyces taught us that antibiotics can be discovered from bacteria. And he wins the Nobel Prize in the 1950s. So this really triggers the so-called golden age of antibiotic discovery in the 50s and 60s. And, from which, the majority of antibiotics was discovered. But today, this particular strain collection contains organisms collected from 40, 50 all the way to the 2010s, so spans more than seven decades.
And think about today, right? Is it possible for you to, say, go to your grandma’s backyard. To collect the diversity that exists in the 1940s? The answer is no! If diversity disappears, it’s never able to be recreated. We can recreate many different things, but we cannot recreate lost diversity. So, sometimes, I think about this strain collection. This is really a museum of diversity. It’s probably a snapshot of all the evolution over the past 70, 80 years. And so, we are not only going to discover new natural products, we also will learn a lot of microbial evolution. We may be able to really see how human civilization is destroying biodiversity over the past 80 years!
DAN: Maybe! How much variation in that collection do you think there was?
BEN: So biodiversity now has been recognized as very unique national resources. So, you may know the so-called Nagoya protocol, which prevents taking bio-resources from the native countries. So it’s becoming very, very hard today to collect microbial diversity, carry them from one country to another. And the strain collection we have here – all of them were collected before 2016, before that true enforcement of the Nagoya protocol. And so, this strain collection from a biodiversity perspective – and I can probably pitch them in two different categories. One is a geographic distribution. So they were collected from 108 countries and regions. So, this is really collected from six of the seven continents and from over 100 countries and regions. So, this would be very, very difficult to do today. And then from the bacterial strains, around a hundred thousand bacteria, and there’s really more than 65, I can say more than 60 different so-called families. So taxonomically speaking, they belong to different species.
DAN: Yeah, yeah.
BEN: And then for fungus, even more. And of the fungus, we have more than 2600 different species. But you have to take the historical perspective, in terms of the biodiversity, because if you are a microbiologist in the 40s, how do you classify a strain? You look at it and this is the 40s – you don’t even know DNA existed! You look at it and this looks to me like, say, micromonospora, so be it, it is Micromonospora! But today, you have so many other say morphological, genetic, you know say sugar, lipid composition – so I think the biodiversity is probably way [more] rich than what we know of based on the record.
DAN: Yeah! I mean any discussion of bacterial speciesism is pretty sketchy anyway.
BEN: So in short I think that this is a really remarkable collection. And the other thing I want to add, you know, we give a loosely-called Pfizer collection, but this is not “Pfizer”, because this also reflects over the past, probably, say, five or six decades, the merger of many pharmaceutical companies. So we got it from Pfizer, but Pfizer is the most current owner of this strain collection. So, this collection is really a collection of half of the bigger pharma in the United States. It goes way back, including Wyeth, Millennium, and SeaTac, Monsanto, even Pharmacia, so there’s a very rich history in terms of scientists over the years that have essentially devoted their life to collect microorganisms all over the world for the sake of discovering biologically active natural products.
DAN: Yeah. Fantastic resource!
BEN: So now we have it under one roof, more importantly. So the other thing I also want to emphasize, I think that’s the part that makes me really excited. You know, you asked, “What are the pitfalls? What are the limitations for natural products, right?” One is we need to develop enabling technology, but not locked in our safe, to make the technology available to the broad – the common scientific community. And that also goes true for the strains. And most owners of the strain, they always prefer to keep it the strain for themselves. So, one of the goals that we at the Scripps Research wanted to do is that, you know, whatever we do, we want to make the strain available to the community, too. So the way I look at it is that, you know, the strain collection, collected all over the world, over the past seven plus decades, right? I view it as like, you have books collected all over the world, written in different times using different languages. So the first thing we want to do is we are also working with JGI is to do genome sequencing, and it’s equivalent to translating all these books into a modern language that everybody can read. So now, if you have, say, a huge library of books, with a unified language, but then you have to have an index. You have to have the computer-searching systems. So one of the enabling technologies we wanted to do is to make this information accessible, so everybody, readers of the library, can come to our databases and look for the specific information they are interested in, in terms of discovering natural products. But, now, if you have a library with the rich information, you have [to have] the best indexes searching for this information. But if they’re not available to the broad audiences, then it will be useless. So the third thing is working, say with JGI, a national lab, or Scripps Research, a nonprofit organization, we are going to make this available to the entire world. And I think that this is not only a game changer for the community, I hope for the outcome which is also going to really facilitate or even accelerate the natural product discovery process. Hopefully that will translate to new medicine.
DAN: Yeah. There’s there’s certainly a lot of work to be done and, you know, with 3 million molecules to be discovered, I don’t think –
BEN: Three million for the bacteria alone! Another 30 million for the fungal!
DAN: Yeah, I don’t think your lab is gonna tackle that single handedly, right? So, yeah, getting the community involved. This is definitely where things need to go.
ALISON: So why didn’t Pfizer hold on to the strain collection? And do this themselves?
BEN: Yeah. So, this has been a major shift. So basically – so the drug discovery you can – to the general public, there’s two major – if I have to think about a scientific discovery, the two major stages. So one is called “discovery”, one is called “development”. So discovery is you find all the innovative leads, and then development is taking the lead into a product. So I think in the past, probably, say, two or three decades, the major change to Big Pharma is Big Pharma becoming so big, and they are very good at moving the lead into a product. But [for] the discovery process, it is really small biotech companies, or, like academic labs, and they are often more nimble and they’re more innovative and so curiosity driven. So the product is not – well, it is our ultimate goal – but it’s not our immediate goal. So I think that for Pfizer to spin off the strain collection and ie the discovery process and to organizations that probably have a better mechanism to make it available to the broader scientific community, is a very great move, but also I have to give Pfizer a lot of credit. It’s really a big contribution from Pfizer’s perspective to the natural product community
ALISON: And I guess that Pfizer and other companies could pick up on the leads that are generated from scientific efforts, now.
BEN: Precisely! Alison, this is a great question. Right? So from Pfizer’s perspective, and not only as I said this biodiversity, if they’re locked in their safe, then nobody will make use of it. So if they make it available, but – They as a for profit organization, they just don’t know. They don’t know how to do this, right? So we can help them to do this. But more importantly, if the community [could] make more discoveries, but ultimately most of the small research labs or nonprofit organizations, we will not be able to make our lead into the product. So we are able to enrich the pipeline, for big pharmas like Pfizer will be able to really now take them into the eventual market.
ALISON: Yeah, yeah, I see how so it’s very synergistic.
BEN: So it’s a win-win proposition for both for the bigger pharmas but also for the scientific research community.
DAN: Do leads have to go back into Pfizer or are there…
BEN: No, no. So there are some subtle kind of … There are subtle obligations but, no. The short answer is no. Scripps Research owns this strain collection. So Pfizer or any other people who discover – the leads that we discover, Pfizer has a right to license and anybody else has the rights to license.
DAN: Sure. Okay, great. Yeah. I mean, that’s important for community uptake. For sure.
DAN: I wanted to know a little bit about your background, what got you into the field of natural product chemistry?
BEN: Yeah. So, I was trained as a plant natural product chemist. When I first started in graduate school, I was working on terpenoids, plant natural products, and what really fascinated me – the pinene – if you go to the forest, you have this very refreshed feeling, right? That’s the flavor of monoterpenoids. The very light natural product that gives you this fresh scent of the fresh air of the forest. So the alpha-pinene… Just [think about] your hands: there’s a right hand and the left hand. And I was told that different regions of alpha-pinene they have a different optical rotation – means right hand or left hand – that fascinated me. Because in man-made alpha-pinene you will have both hands, but nature will only make one of the two hands.
So that was my introduction, that nature was able to do something very unique. Now if you think about this, as a natural product chemist, immediately you’re asking the question why in nature are you making only one of the two hands. So that’s a question about biosynthesis. So, I took this question in mind, and come to the United States and was working on natural product biosynthesis, ie to ask the question of how nature’s making compounds. Now that was in a time when we don’t know how to sequence DNA. So, we were essentially using brute force to isolate the proteins. The proteins are the biological version of the catalyst to catalyze the production of the natural product.
So that was my PhD study, and then towards the end of my PhD study, then recombinant DNA technology was becoming available. So I saw this as a great opportunity. So I wanted to learn DNA, recombinant DNA. So when I first started work, my independent work in and in 1995, I thought. I’m going to combine natural products isolation, enzymology, and writeable recombinant DNA technology to study natural product biosynthesis. No one could predict it’s only going to take five years before we have the first genome. So in 2000 genome sequence became available. That changes everything!
So, that leads me today to, essentially, to combine all the emerging technology. And we are still asking the fundamental question, how is Mother Nature making natural products? But we are no longer using, say, my mentor’s generation relying on brute force of conventional chemistry, biochemistry, but instead, we’re using all the emerging technology. So, we are not only trying to understand how nature is making compounds, but we are also applying CRISPR technology, anything as genome editing, genome engineering, to make something even better and making more of it, and making designed tailored molecules for our specific applications.
DAN: Yeah, that that broad use of many technologies is sort of one of the hallmarks of the field of natural products and secondary metabolism. So it’s always fun to sort of incorporate new technologies and things because it just opens up new doors. Yeah. Yeah, your origin sounds somewhat similar to mine. I was talking to an organic chemistry professor about some reaction and why the stereochemistry was that way. And he told me, it’s enzymes. It’s magic.
BEN: Yeah, right.
DAN: I thought that was the worst explanation a scientist could give and got inspired.
BEN: Well, the other thing I really think about it in earlier days, right, when I was in graduate school, and we have a department, we have divisions, we have fields called organic chemistry, right? But today in many places, like, say, if you go to Scripps Research, we don’t have departments anymore, it’s just science. So the boundary of traditional disciplines clearly is disappearing. And it is the fundamental question we are asking. Right. And we’re still asking probably similar fundamental questions, but [using] whatever method that works, and you’re going to apply for it. The other thing I also think is very fascinating is that with the genomics, with the bioinformatics and with the bigger data, another area I see a great potential will be machine learning and artificial intelligence. I would view that will be the next frontier. Yeah, because if we do all this genome sequencing, as we would like to do, for the hundred thousand bacterial strains, and hundred thousand fungal strains, that will generate a massive amount of data that current technology has never encountered in the past. That’s even for places like JGI.
DAN: No, yeah, for sure. I mean, I can look at a biosynthetic cluster and sort of figure out what’s going on. But I can’t look at, you know, 3 million biosynthetic clusters.
BEN: So that would put – analyze what natural products taught us in the past. And the natural products taught us fundamental structures, fundamental biology, fundamental medicine. And this is mass data, the huge databases, is probably also going to inspire us to really apply some machine learning artificial intelligence into the data mining and in return to help us discover more natural products. So that will be an iteration of tech that takes us to the next stage of natural product biosynthesis, discovery, or natural product-based drug discovery, in general.
DAN: All right, that sounds like a great place to stop. So Ben, thank you so much for talking with us today. Thank you for chairing this great conference. And let’s go look at some posters.
BEN: Great! Thank you very much for having me. This is fantastic!
ALISON: Yeah. Thanks so much, Ben!
DAN: I’m Dan Udwary, and you’ve been listening to Natural Prodcast, a podcast produced by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility located at Lawrence Berkeley National Lab. You can find links to transcripts, more information on this episode, and our other episodes at naturalprodcast.com
Special thanks, as always, to my co-host, Alison Takemura. <woohoo> If you like Alison, and want to hear more science from her, check out her podcast, Genome Insider. She talks to lots of great scientists outside of secondary metabolism, and if you like what we’re doing here, you’ll probably enjoy Genome Insider too. So, check it out.
My intro and outro music are by Jahzzar.
Please help spread the word by leaving a review of Natural Prodcast on Apple podcasts, Google, Spotify, or wherever you got the podcast. If you have a question, or want to give us feedback, tweet us @JGI, or to me @danudwary. If you want to record and send us a question that we might play on air, email us at jgi-comms@lbl.gov. And because we’re a User Facility, if you’re interested in partnering with us, we want to hear from you! We have projects in genome sequencing, DNA synthesis, transcriptomics, metabolomics, and natural products in plants, fungi, and microorganisms. If you want to collaborate, let us know! Find out more at jgi.doe.gov/user-programs.
Thanks, and see you next time!
Transcribed in part by https://otter.ai