For the first time, a team analyzes the transcriptomes of a lichen fungus and alga to understand their partnership more clearly.
The Science
The humble lichen is a superorganism: one being that is actually comprised of two (or more) participants. One is a fungus (usually belonging to the ascomycetes, one of the two main branches of fungi); another is a photosynthetic microbe, tucked in the fungus’ tissue. If you’re a Marvel Comics fan, you might liken the symbiotic relationship to that of Eddie Brock and Venom, an alien “symbiote” that colonizes Brock and gives him super strength. For lichen, the partnership is more commonplace — though perhaps no less dramatic. In lichen, the photosynthetic partner, either a eukaryotic microalga or a cyanobacteria, is known as the “photobiont.” Photobiont and fungus (mycobiont) live intertwined, with the photobiont transferring part of its photosynthetically fixed carbon to the mycobiont. However, despite a century-and-a-half of lichen research, many of the symbiosis’ details remain unclear. For the first time, a team has analyzed in parallel the genomes and transcriptomes of both partners to better understand lichen.
The Impact
Lichens are a model for inter-kingdom communication and cooperation. Their partnership is widespread and cosmopolitan: more than 20,000 ascomycete species and hundreds of microalgae are able to partner to form lichens. Studying the symbiosis by investigating a lichen’s genomes and transcriptomes could help inform efforts at bioengineering symbiotic microbial consortia for bioenergy crops. And because lichens are dominant carbon and nitrogen fixers in alpine and high latitude ecosystems, deciphering their internal molecular mechanisms can help researchers better model global carbon and nitrogen cycling.
Summary
To shed light on lichen, an international team led by Daniele Armaleo, a biologist at Duke University, investigated Gray’s Cup lichen (Cladonia grayi). The lichen’s primary components are the fungus it’s named after, Cladonia grayi, and the microalga, Asterochloris glomerata.
To see how gene expression changes when living together versus apart, the team grew the fungus and alga, either separately or together, on nitrocellulose filters in Petri dishes. The scientists also sequenced the fungus and photobiont’s genomes and compared them to close relatives who don’t engage in the lichen lifestyle. The team’s results appear in BMC Genomics.
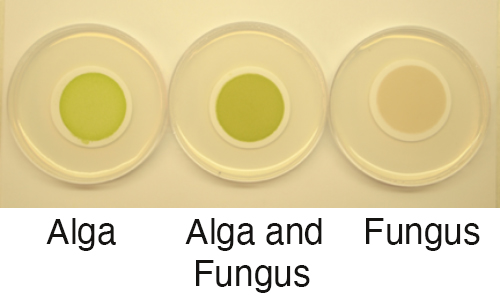
Cultures of fungus C. grayi and microalga A. glomerata grown individually and together on filters in the lab. (Courtesy of Daniele Armaleo)
To explore the genomes, Armaleo and colleagues worked with Igor Grigoriev, the Fungal & Algal Program lead at the US Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility. Grigoriev and his team at the JGI contributed to annotation of the genomes, available online at PhycoCosm (for the alga genome) and MycoCosm (for the fungus genome) to aid analysis.
Many of the results were unexpected. For example, lichenologists generally anticipated that in this symbiosis going back hundreds of millions of years, accidental gene swaps via horizontal gene transfer would have occurred between the partners. Yet the team found none. Instead, the partners stubbornly maintained complete autonomy; they can be isolated and grown separately from each other.
A possible explanation, according to the authors, lies in their finding that each lichen partner can sexually reproduce: genetic recombination during sex could increase the fitness of both partners. While it’s well documented that the lichen fungus can reproduce sexually by releasing spores, the alga was generally thought to develop clonally. Yet its genome revealed that the alga has all the genes needed for meiosis, and thus can rapidly create genetic variation in its progeny.
The study’s transcriptomic data also identified many genes potentially important for the symbiosis, including those involved in carbon and nitrogen exchange between the lichen partners.
This genomic and transcriptomic analysis has opened the door for more incisive investigations into how lichens function. One of their abilities that’s now getting more scrutiny is how lichen is able to survive drying out to a potato-chip level of crispiness, only to revive in mere seconds or minutes — a trait that could help improve crop drought resilience. It’s a superpower even Marvel’s Brock/Venom would envy.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
PI Contact
Daniele Armaleo, Ph.D.
Duke University
darmaleo@duke.edu
Funding:
This work was supported by an intramural seed grant (IGSP 2008001) to DA, FL, and FD from the Duke Institute of Genomic Sciences and Policy (for DNA sequencing and assembly); by a three-year subcontract to DA, FL, and FD (for bioinformatics) from a DOE grant No 112442 to Pacific Northwest National Laboratory; and by a DOE-Biological Environmental Research grant No FWP 61327 to FRC and PL (for RNAseq). The work at the Joint Genome Institute is supported by the U.S. DOE Office of Science under Contract No. DE-AC02-05CH11231. The work of MP, DL and SSM was supported by the U.S. DOE Office of Science, Office of Biological and Environmental Research program under Award No DE-FC02-02ER63421. The meiosis gene identification was performed in the context of work supported by the National Science Foundation under Grant No. 1011101 to E.S. and J.M.L.
Publication:
- Armaleo D et al. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterochloris glomerata. BMC Genomics. 2019 July. doi: 1186/s12864-019-5629-x
Related Links:
By: Alison F. Takemura
