Machine learning approach significantly expands inovirus diversity.
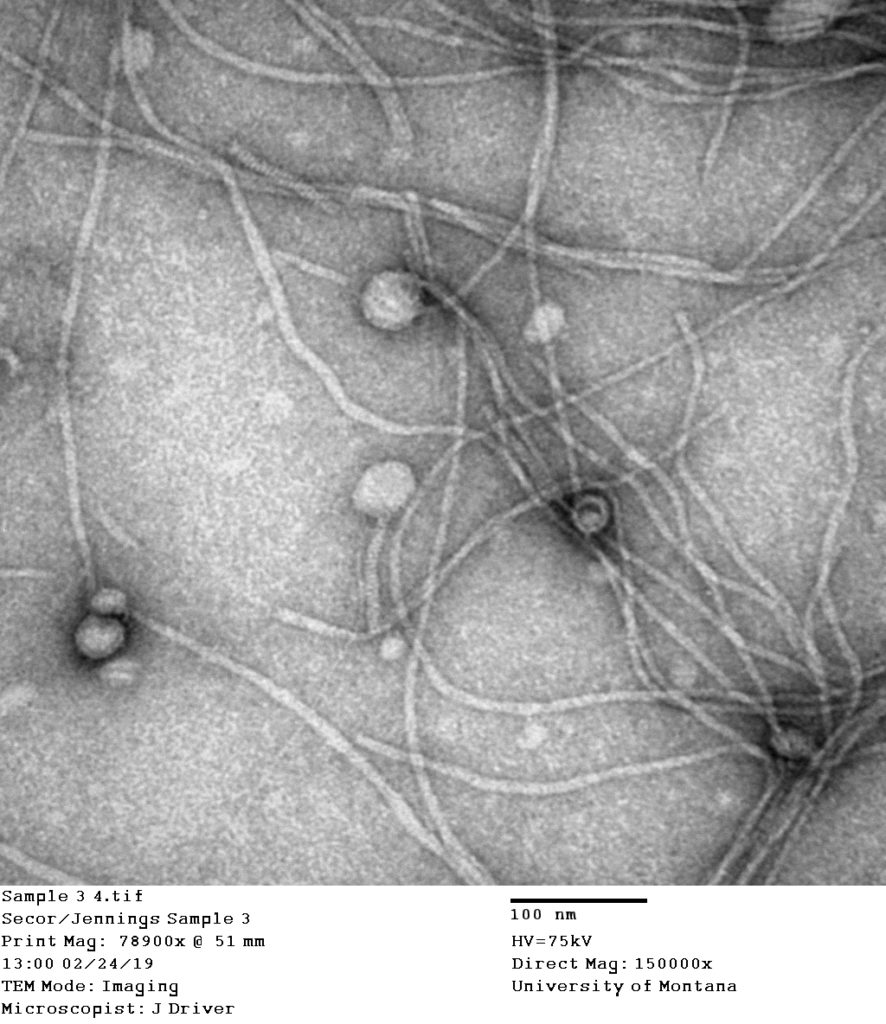
EM image of Pseudomonas phage Pf, an inovirus infecting Pseudomonas hosts. Inovirus capsids are long flexible filaments visible here after sample concentration and precipitation. (Courtesy of J. Driver and P. Secor, University of Montana)
To answer the question, “Where’s Waldo?” readers need to look for a number of distinguishing features. Several characters may be spotted with a striped scarf, striped hat, round-rimmed glasses, or a cane, but only Waldo will have all of these features.
As described July 22, 2019, in Nature Microbiology, a team led by scientists at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility, developed an algorithm that a computer could use to conduct a similar type of search in microbial and metagenomic databases. In this case, the machine “learned” to identify a certain type of bacterial viruses or phages called inoviruses, which are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group.
Training the Search Tool
Inoviruses are stealth agents that can enter and exit through the cell membrane without lysing the bacterial host. They can also influence their host’s growth and pathogenicity, in turn affecting the microbe’s own eukaryote host. As their small genomes can be easily manipulated through genetic engineering, inoviruses are used for several biotechnological applications, most notably, phage display. The search tool Roux and his colleagues developed first worked on a reference dataset that included genome sequences known to be affiliated with the Inoviridae. “What we’re really doing is looking for a particular gene found in all inoviruses, and then checking the surrounding genes,” he said. “If these genes are similar in size and function to those in typical bacterial or archaeal genomes, the sequence is most likely not an inovirus. But if these nearby genes are both short and novel, then that’s a very good indicator that it is a genuine inovirus.”
After Roux manually curated the results and refined the algorithm, the search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. “These genomes are so special, regular search methods don’t work,” said Roux. “The machine learning approach allows you to quickly scale up once you’ve found the right features that you can use to identify the inoviruses.”
Overhauling the Perception of Inovirus Diversity
The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world. By the numbers, the new approach detected inoviruses in 3,609 (6 percent) of the 56,868 microbial genomes and 2,249 of the 6,412 (35 percent) metagenomes mined for this study. “We’re simply getting much better at seeing them, which means we can now study their biology much more meaningfully,” Roux noted of the result.
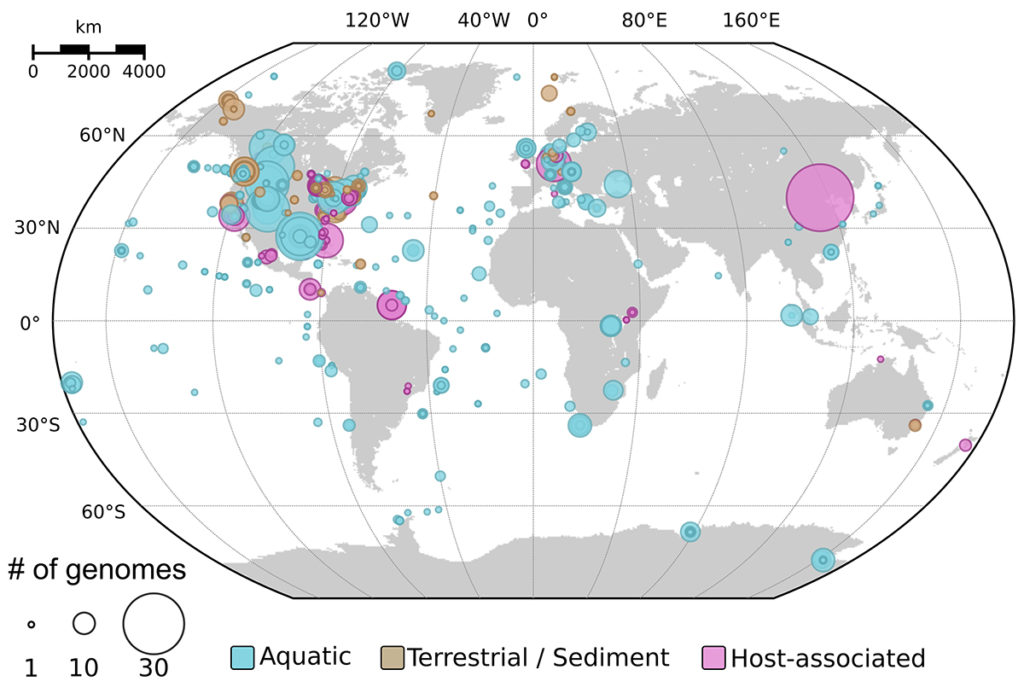
Detection of inoviruses across ecosystems. The map displays the location and type of samples from which new inovirus sequences were identified as part of the global metagenome search. (Simon Roux)
“It troubled me for a long time that we had only a handful of representatives of this virus group,” said virologist Mart Krupovic of the Institut Pasteur, one of the study co-authors and an expert on inoviruses. “The result of this hidden diversity of inoviruses now overhauls our perception of this virus group – from minor curiosities they become a prominent component of the prokaryotic virome associated with nearly all bacterial phyla across virtually every ecosystem.”
By significantly expanding the known diversity of these viruses, genomic analyses led the team to propose that the Inoviridae should be classified as an order of viruses, with six families. Additionally, the team uncovered a range of genetic diversity among inoviruses with more than 3,400 different proteins, many linked to key functions such as virion structure and extrusion, and DNA replication and integration. The researchers also learned how an inovirus’ strategy of integrating itself within a host can lead to beneficial or antagonistic interactions with other co-infecting phages and with the host’s CRISPR-Cas immunity systems.
Countering Co-Infections, Ensuring Host Survival
Many bacteria have CRISPR-Cas systems that incorporate short sequences from infecting viruses and phages to help the bacterial host resist foreign genetic elements. In some cases, Roux and his colleagues found that the inoviruses were being targeted by their hosts’ own CRISPR-Cas systems, termed “self-targeting,” and yet still survived. The persistence of these “self-targeting” inoviruses suggested they had found a way to deactivate the CRISPR-Cas systems, and led the researchers to predict the presence of anti-CRISPRs, recently discovered inhibitors of bacterial CRISPR-Cas systems that have no conserved structural motifs or domain architectures.
“Anti-CRISPRs are important from the standpoint of phage-bacterial coevolution and are also useful tools in CRISPR-Cas applications, but we are limited in our predictive methods to discover new anti-CRISPRs,” said Adair Borges, a graduate student in bacterial immunologist Joe Bondy-Denomy’s lab at the University of California, San Francisco. Both are co-authors on the study. “By finding a new anti-CRISPR locus, in an inovirus for example, we would be able to discover all the new anti-CRISPRs that are associated with that genetic neighborhood. So anti-CRISPR loci are powerful discovery tools, by finding even one new anti-CRISPR locus, you are unlocking many new anti-CRISPRs.”
Borges worked with Roux and found that the inoviruses don’t need to make their own anti-CRISPRs. Instead, she said, the inoviruses she studied “piggyback” off the anti-CRISPRs made by the co-infecting phages in the same host cell, relying on their shared desire to avoid CRISPR-Cas immunity. In addition, Borges also showed that inoviruses might prevent new phages from infecting a cell in which the inovirus has established itself through a process called superinfection exclusion, which is another way by which they can help their host survive.
“It is an exciting time to be studying filamentous phages!” said Krupovic. “We can now start inquiring into their impact on microbial communities in the environment and also those associated with humans.”
The work also used resources of the National Energy Research Scientific Computing Center, another DOE Office of Science user facility.
Publication: Roux S et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nature Microbiology. 2019 July 22. doi: 10.1038/s41564-019-0510-x
Nature Microbiology Research Community “Behind the Paper” blog: Simon Roux on Expanding inovirus diversity: the rise of the small and inconspicuous.
Byline: Massie Santos Ballon