Every summer, the flagship JGI-UC Merced Internship Program invites undergraduate and graduate students to engage in real, impactful research projects with JGI mentors. In this episode, hear two interns from the 2021 cohort describe their deep dives into genomics, computational tools, and big data.
The Joint Genome Institute presents the Genome Insider podcast.
Transcript of the episode
ALISON: Hey! I’m Alison Takemura, and this is Genome Insider, a podcast of the US Department of Energy Joint Genome Institute, or JGI. For this episode, former JGI intern, now staff science communication colleague Ashleigh Papp will be joining me as co-host!
ASHLEIGH: Hi!! It’s great to be back. In this episode, we’re going to be talking about an internship program at JGI.
ALISON: This program started in 2014, and it’s a partnership between JGI and UC Merced. Each summer, JGI scientists take UC Merced students under their wing! The interns are usually about equal numbers graduate students and undergraduate students, and their backgrounds range from biology and physics to computer science and engineering. In the past, the interns have had hands-on experiences in the lab. But with the COVID-19 pandemic, the last two summers have been virtual, making all the internships, for now, focused on bioinformatics.
The 2021 JGI-UC Merced cohort, made up of 13 interns and their mentors, came together in July 2021 to virtually present their summer research projects. (Ashleigh Papp)
ASHLEIGH: One of the coolest things about this program, I think, is that the students get to decide which project and mentor they work with throughout the summer. Mentors from JGI submit project proposals that each student, once they’ve been selected, reviews and ranks according to their interests and goals. Some of the interns choose projects that align with their studies, giving them a deep dive into their budding field of expertise. While others, choose a project that’s completely outside of their comfort zone in an effort to broaden their skillset.
ALISON: On top of their project work, the interns also tune in to professional development activities, fun virtual lab-wide events, and plenty of networking dates, too. Overall, the goal of this program is to introduce genomic research to the next generation of scientists.
ASHLEIGH: And our mentors, and JGI as a whole, learn a lot along the way! Many JGI scientists who have participated in the program as mentors have shared how impactful that fresh, intern perspective has been to help further their research. Overall, the program has been a win-win.
ALISON: In this episode, we’ll hear from two UC Merced interns and their mentors, all of whom we interviewed this summer.
ASHLEIGH: First up is Rahul Ravi, now a third year undergraduate student at UC Merced.
RAHUL: I’m a cognitive science major. So my study is the study of human behavior. And I’ve combined that with applied math.
ALISON: But instead of studying about humans and their brains, his project this summer focused on better understanding a type of grass and its genome. This grass is Brachypodium distachyon, or just Brachypodium, for short. So you can get a picture of this plant, it’s a vivid green with just a touch of blue. And it’s name, “Brachypodium,” means ‘short, little foot.’ And it lives up to that title: It grows only to about 5 to 8 inches tall. Honestly, it’s very unprepossessing; it looks like something you’d pass on the sidewalk without a second glance.
ASHLEIGH: But, it’s an important plant because we like to use it as a model system. It has a compact genome and shorter life-cycle than the more complex bioenergy grasses that the DOE is targeting. Think, things like switchgrass, sorghum, and Miscanthus — we’ve done episodes on both switchgrass and sorghum, so check ‘em out if you’re curious!
Brachypodium is easier to study, so we can try to learn things about the biology of grasses first in Brachypodium, and then extend those findings to other bioenergy grasses.
RAHUL: So what I’m working with is discovering genetic variations, because this is very important in both the medicine and agricultural field.
ASHLEIGH: Rahul’s project was to take the Brachypodium genomes that the JGI has sequenced and look at all the genetic variation within them. JGI scientists want to understand what makes these individual Brachypodium genome sequences different. Because, plants, like people, even if they belong to the same species, are not all the same.
ALISON: Rahul and Albert worked with four Brachypodium genomes. The first one sequenced, in genome science parlance, is called the reference. Now it’s very hard to get plant genomes to be 100% complete, so all of these genome sequences hit around the 98% mark. The genomes are broken into fragments, with gaps of unknown stuff between long stretches of known sequence.
ASHLEIGH: So this is what scientists have to work with when they’re comparing genome sequences. Rahul’s project was to compare these Brachypodium genomes using some novel approaches, because, as his mentor, JGI Plant Program data scientist, Guohong Albert Wu, explains, the historical approach just wasn’t up to snuff.
ALBERT: The old approach is based on a single reference genome, and then map the short DNA fragments from other plants to the reference genome. And identify whether there’s variation, for example, for a particular gene, was this particular amino acid, whether it’s changed or not, but this approach has a very strong limitation. That is, if a gene is not present in the reference genome. So it will be missed in this approach.
ALISON: Rahul likens this process of looking for and noticing these differences between genomes to walking down a grocery store’s cereal aisle.
RAHUL: You might find a few cereals that look the same, but they are a bit different, like some might be sugar-free, some might be keto-friendly. And similarly to that, when looking at these Brachypodium genes they have slight variations that we need to be mindful of.
ALISON: Because, as Rahul puts it..
RAHUL: You might find a cereal that has sugar but the reference cereal has no sugar.
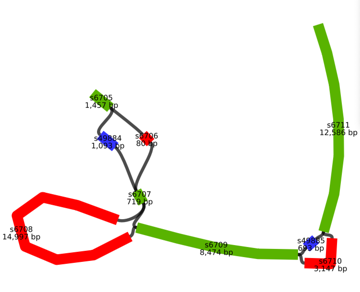
ASHLEIGH: Rahul’s project this summer has been to compare Brachypodium genomes using new software tools — called Minigraph and VG, which stands for Variation Graphs. These tools don’t exclude any genes or partial gene sequences and allow users to visualize the genome comparisons. Like their names suggest, they render a genome comparison in the form of a graph.
ALISON: This kind of graph comes from a particular branch of mathematics, and so it looks different than your typical graph. These graphs are like networks, made up of points of interest, called nodes, and lines that connect them. To see examples, check out the transcript for the episode online! OK, so how do these graphs look when we’re talking about genome comparisons? In this case, the points of interest are stretches of shared or unique DNA sequence, and the lines are how they’re connected. To walk us through an example, here’s Albert.
ALBERT: Suppose there are two individuals share the sequence, then it’s just one node. And then at a particular gene, these two individuals diverge, meaning the genes are quite different. Then you start splitting into two paths. So one individual take on each path. Then they merge again as the sequence from the two individual converge.
ASHLEIGH: It’s kind of like looking at a trail map when you go hiking. One path might split from another before meeting up with it again. When comparing genomes, it’s helpful for scientists to see these paths and divergences.
ALBERT: People, for example, may be interested in a particular gene they have been studying for years, and they want to see what’s the variation of this gene among a given population. Then this visualization will allow them to zoom in this gene.
ALISON: ..and see if maybe there are some particular sections of the gene that take different paths in sequence space. That’s a really helpful insight for scientists.
RAHUL: It just makes it easier for them to analyze and compare the differences between sequences and really find what part of the DNA is resulting in these changes.
ASHLEIGH: What did Rahul get out of this summer internship? New skills and a broader perspective on science.
RAHUL: This was my first formal research internship. And so I think that it gave me some valuable insight into how research is conducted at a national laboratory. And also learn how to write scripts and work with the Unix command line, and Unix text editors and work with big data files and write scripts that can read and write big data files.
I’ve never really worked with big data before. And when working on projects, just by myself, I don’t have the resources like JGI does to sequence DNA and work with data files of like gigabytes and terabytes, so that was really interesting to work with.
ALISON: Just for comparison, one terabyte of data is the equivalent of streaming something like 400 movies in HD. That’s the scale of this big data. Back to Rahul — working with the JGI, Rahul noticed that there’s a different level of contribution that his work can make.
RAHUL: I felt like I was pushing the needle further for science, and maybe making some contributions that could be carried out further, whereas when I just work on personal projects, it’s more of just to make my life easier, like make some scripts to run my applications in the morning, so I don’t have to, like manually click on the buttons or something like that.
ASHLEIGH: Rahul’s coding ability was a big help to Albert, too.
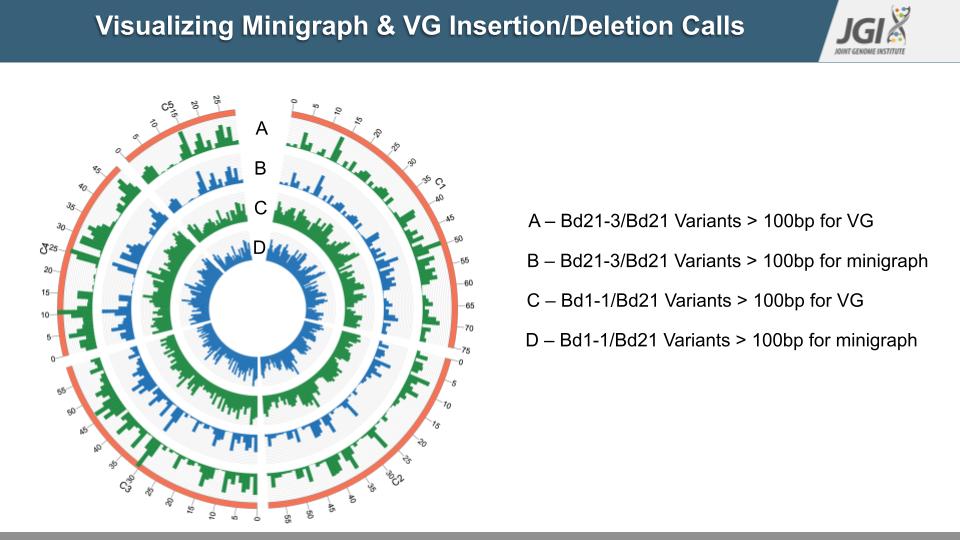
A slide from Rahul’s final presentation of his project work with Albert, depicting the visualizations possible with Minigraph and VG. (Rahul Ravi and Albert Wu)
ALBERT: I’m very impressed by Rahul’s coding skill. He keep writing new Python scripts to solve different gene analysis problems we encounter. So Rahul’s very efficient.
Another impressive thing about Rahul was his self-learning capability.
ALISON: For example, if there was a particular data processing problem that he and Albert wanted to address,
ALBERT: ..he would quickly find the best available software package maybe already somewhere on the internet. Or if you want to write a new script, and need to learn some new way of coding, he can quickly identify the right syntax by going to the internet. Basically, yeah, it’s all different aspects of his strong skill in coding and solving problems. That’s a very strong asset. Not only for this summer, but in his future endeavors.
ASHLEIGH: Rahul, being a cognitive science major, had never worked with plant genomes before. But our next intern tackled a subject that’s a little closer to home: the vast, crazy world of viruses!
CLARENCE: I’m Clarence Le. I’m a UCM-JGI graduate intern for the 2021 summer and I’m working on a project to identify new and novel virus groups in environmental samples.
ALISON: With Clarence being a graduate student intern, he came in with some prior experience studying human immunodeficiency virus, or HIV. And that made a nice springboard into his JGI internship project to mine samples sequenced by the JGI in order to look for new kinds of viruses.

Clarence Le, graduate student at UC Merced. (Courtesy of Clarence Le)
CLARENCE: And so for my project, we’re specifically interested in RNA viruses.
ASHLEIGH: RNA stands for ribonucleic acid, meaning that these types of viruses don’t have DNA, but instead carry their genetic info as RNA. Examples of RNA viruses include the common flu, HIV, polio, and you guessed it — SARS-CoV-2, the virus that causes COVID-19.
ALISON: All those viruses affect humans. But RNA viruses can infect hosts of all shapes and sizes.
CLARENCE: The most studied and known RNA viruses are found in plants and animals.
ASHLEIGH: But these viruses can also infect teeny, tiny living organisms that we can’t see with the naked eye.
CLARENCE: And so that’s kind of like a level that we were interested in going into.
ALISON: Clarence had his eyes on RNA viruses infecting microorganisms. For his project, he worked with Frederik Schulz, a research scientist at JGI who leads the New Lineages of Life group.
FREDERIK: Hi, my name is Frederik Schulz and I’m a scientist at the JGI and I’m leading our efforts towards discovering new lineages of life. That includes new bacteria, archaea, and eukaryotes, and also viruses, and everything else unusual that we find in the data. When we look in the data, and we find something really novel, it’s very different from everything we know already, and sometimes, I mean, we don’t know what it is yet.
ASHLEIGH: And, as Frederik quickly points out …
FREDERIK: There are a lot of strange things in the data often. Some of the RNA viruses are, in fact, among those very strange sequences that we find. That’s something this project that we maybe can shed more light on.
ALISON: We know that viruses infect their hosts, and usually that leads to something bad happening. We get sick, plants or cells die, that kind of thing. But according to Frederik, we’re also interested in those genes in RNA viruses that might potentially help a host.

Frederik Schulz, research scientist at JGI who leads the New Lineages of Life group. (Courtesy of Frederik Schulz)
FREDERIK: We’re also interested in if there are any other genes in this RNA viruses that potentially complement host metabolism like rewire kind of the host cell. And we, we know this from bacteriophages, and also from giant viruses that they encode for many different genes, like transporters, and genes that are potentially involved in photosynthesis, other genes like for fermentation processes, and so on. There’s a long list of genes that viruses may encode and potentially get readily into the host, where the host then can do things they couldn’t do before. Ultimately, maybe competing better with other host populations that are infected by other viruses. So the virus wants to out compete. These kind of things, they’re completely not found yet in RNA viruses.
ASHLEIGH: So Frederik and Clarence set out to find new viruses. Because maybe they’re not just making organisms sick.
ALISON: To search for RNA viruses in the data, Clarence and Frederik used a characteristic protein — or feature – of RNA viruses.
CLARENCE: So we’re looking to all the sequences and determine like, which ones are we going to include in our analysis. And so the feature that we’re looking at was called RDRP. And so that stands for RNA-dependent RNA polymerase.
FREDERIK: So this RDRP proteins, they can be quite divergent between the different groups of RNA viruses that we know. And so we try to learn from these sequences and from the protein structure to find ways to also discover viral groups that we haven’t seen yet. And because in theory, every RNA virus should have this RDRP. And so we basically build models, and we use those models to screen the environmental sequence data to find novel RNA viruses.
ASHLEIGH: It’s worth mentioning that this type of work, which relies almost entirely on models, is based on what we already know. So there’s a bit of a bias … which means you have to take this type of research with a grain of salt.
ALISON: But it gives Frederik and Clarence a place to start. What did Clarence do next?
ASHLEIGH: Clarence used NERSC, which stands for National Energy Research Scientific Computing Center. It’s one of DOE’s supercomputing centers, operated by Berkeley Lab.
Along with this center’s computing strength, Clarence used specially-designed code to worked through the massive data sets and find new viral sequences. He also used a phylogenetic tree to map out what species were there.
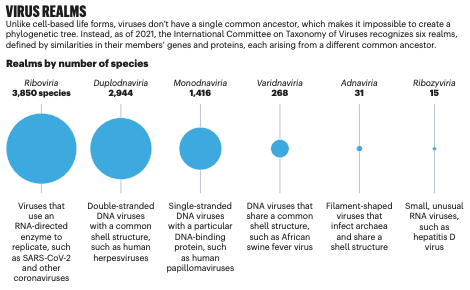
A summary of research on the origins and classification of viruses. (Figure from Dance, A. The incredible diversity of viruses. Nature. 2021 Jul 1. https://doi.org/10.1038/d41586-021-01749-7)
CLARENCE: We’re basically trying to see like, when we put them on to a phylogenetic tree representation, like do they fall into groups that we already know about? Or do these new viruses kind of form their own new groups, new clades.
ALISON: I.e., groups of viruses that were totally unknown before.
CLARENCE: And so that’s kind of what we’re starting to see.
ASHLEIGH: One of the great things about this internship program at JGI is that it can introduce new topics and new ideas to students. Here’s Clarence:
CLARENCE: Before I did this project, the word like “genomics,” like I’d have friends that told me they were doing genomics research, or like, dealing with like sequences, I didn’t know what that entailed until this project. And so this project really made me appreciate like the field of like genomics. Using like such a simple thing like, a genomic sequence, painting a larger picture with it, and kind of like, being able to answer actual scientific questions with such a, such a simple looking text file that’s on your computer.
ALISON: And another plus about internships is that they help students get out of the orderly classroom environment where, sometimes, the messier parts of research get left out.
CLARENCE: Through this process, I do have a newfound appreciation for, like, data cleaning or data wrangling. I was used to getting very clean data sheets, that were just columns and rows, and then I can just have fun with it. And then this made me appreciate the process of actually, like, accessing public databases, cleaning up data down to the titles, and then like parsing things and just going through the process of actually sifting out like the gold from all the rocks.
ASHLEIGH: While the interns have had an enriching experience, the JGI also really benefits. Here’s the JGI Director Nigel Mouncey.
NIGEL: Having the opportunity through this internship program to work with this next generation of scientists, is incredibly rewarding, I hope for very much for the scientists, but also for our folks too. We’ve had some wonderful interns over the years that we’ve had this program. And, I’d like to think that as much as they are learning from us, we’re learning from them.
ALISON: How is that? In part, it’s because of the diverse backgrounds of the interns.
NIGEL: We value highly diversity in biology. And that’s not just the types of samples from the diverse environments or the diversity of life that we are investigating, but the diversity of those people that we work with, whether it’s at the JGI, or across the scientific community. And especially important to this is the training and development of the next generation of genome scientists that are really going to continue this journey in exploring this incredible diversity, of being able to unravel the layers of complexity. So that, we can increase our knowledge of biology.
ASHLEIGH: Nigel sees training future scientists as part of JGI’s mission.
NIGEL: I think it is one of our responsibilities in science, to be able to help educate others, and, to hopefully, be able to hire some of those people, as the next JGI scientists.
ALISON: This episode was directed and produced by Ashleigh Papp and me, Alison Takemura, with editorial and technical assistance from Massie Ballon and David Gilbert.
ASHLEIGH: Genome Insider is a production of the Joint Genome Institute, a user facility of the US Department of Energy Office of Science. JGI is located at Lawrence Berkeley National Lab in beautiful Berkeley, California.
ALISON: A huge thanks to these past JGI interns and scientists in training: Rahul Ravi and Clarence Le at UC Merced. Thanks, too, to their invaluable mentors: JGI Plant Program data scientist, Guohong Albert Wu, and research scientist Frederik Schulz who leads the JGI New Lineages of Life group. And a big thanks to JGI Director Nigel Mouncey for talking about training the next generation workforce.
ASHLEIGH: If you enjoyed Genome Insider and want to help others find us, leave us a review on Apple Podcasts, Spotify, or wherever you get your podcasts. If you have a question or want to give us feedback, Tweet us @JGI, or record a voice memo and email us at JGI dash comms at L-B-L.gov. That’s jgi dash c-o-m-m-s at l-b-l dot g-o-v.
ALISON: And because we’re a user facility, if you’re interested in partnering with us, we want to hear from you! We have projects in genome sequencing, synthesis, transcriptomics, metabolomics, and natural products in plants, fungi, algae, and microorganisms.
ASHLEIGH: If you want to collaborate, let us know! Find out more at jgi.doe.gov forward slash user dash programs.
ALISON: And if you’re interested in hearing about cutting edge research in secondary metabolites, also known as natural products, then check out JGI’s other podcast, Natural Prodcast. It’s hosted by Dan Udwary and me.
That’s it for now. See ya next time!
Additional information related to the episode:
- JGI-UC Merced program:
- JGI-UC Merced Interns Share Their 2021 Experiences
- UC Merced Interns Reflect on Their JGI Summer 2020 Projects
- Toasting to the 2020 JGI-UC Merced Summer Internship Program
- Scaling Up Existing Skill Sets
- High Five: Celebrating the JGI-UC Merced Internship Program
- Access to Rock Star Scientists
- 2017 Interns: A Social Science Experience
- JGI Interns at UC Merced Earn CAMP Merit Awards
- 2015 JGI Interns Share Their Summer Experiences
- Training the Next Generation of Talent
- JGI Brachypodium Resources
- 2021 New Lineages of Life Symposium
- Our contact info:
- Twitter: @JGI
- Email: jgi-comms at lbl dot gov