In late June, Kathleen Lail, a member of JGI’s Sample Management Group, attended a conference in Florida to discuss a JGI pilot project involving soil sampling in the Florida Everglades to help train the next generation of scientists. She shares her thoughts about the experience below.

Boca Raton Community High School students (in alphabetical order): Deniz Caglayan, Claire Hagan, Ralph Jeanty, Alexander Klimczak, Sydney Levy, Angel Millard-Bruzos, David Prentice, Richard Roehm, Mulan Yin, with teacher Jonathan Benskin (second from left) presented at the E4 community-building workshop. Also present: David Blockstein (center), one of the Association for Environmental Studies and Sciences (AESS) founders and workshop session leader; and Kathleen Lail (leftmost) of the JGI.
In the summer of 2018, Jonathan Benskin was preparing to teach a second year A-Level Advanced International Certificate of Education (AICE) Biology class at Florida’s Boca Raton Community High School. The course had a defined set of objectives, and started with an image of a spoonful of dirt with the question: Do you want to find out what is in this soil?
Jonathan had been reaching out to many researchers with the hope of getting support for a research project for his class of 18 juniors. He got turned down consistently, with one of his favorite answers being, “It would take a team of Ph.D.’s over a year to do a project at this capacity.”
Jonathan’s thought to himself was, “Game on.”
Then Jonathan’s email request reached Emiley Eloe-Fadrosh, the Metagenome Program Head at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility. She took special interest in it because she credits her high school teacher with inspiring her to get into science and wanted to pay it forward.
Defining a Diverse Research Topic
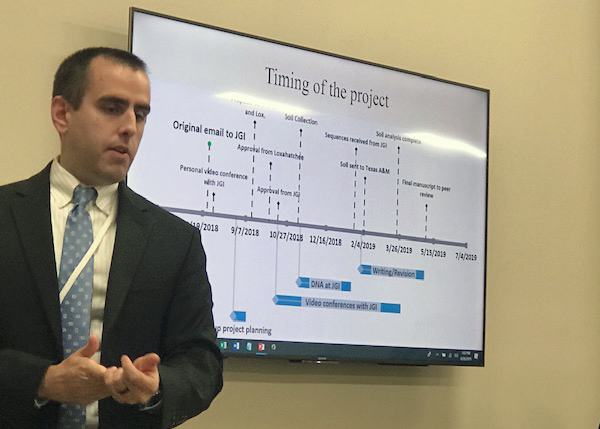
After a video conference interview between the class and the JGI, the students found out that their project was approved and was being funded under the JGI Director’s Science Program. The class did not know at the time that the project had already received enthusiastic support from the JGI Director, Nigel Mouncey.
Jonathan credits his intrinsically motivated students with fully defining the project, but it also took a very motivated teacher willing to work and learn in a discipline without the expertise, along with the students. The project provided the students the freedom to define the specifics. It took the coming together of many unlikely pieces across the United States to enable the class to perform research that would have been difficult, if not impossible, without such a serendipitous partnership.
The class decided to do an environmental microbial community (metagenomic) soil study in an untouched region of the Everglades, the Loxahatchee National Wildlife Preserve. They chose this location because much is unknown about the diversity of organisms (taxonomy) and particularly those in processing (metabolizing) methane within the Florida Everglades ecosystem.
There are no roads or real human traffic through this remote part of the Everglades. The students had to come up with an extraction procedure for the soil and acquire the permits to do so. They used images from Google maps to plan their route, and had to adapt when they noticed while collecting the samples on canoes just how much change had occurred in this region since the images used to map their routes had been taken.
Sifting through 16 Trillion Bases of DNA
Taking into consideration some complicated environmental challenges for collecting these soil samples, the students made coring devices, then planned and collected from 4 sites with 5 replicates. They extracted the DNA from the soil with Qiagen DNeasy PowerSoil Kits and sent the samples to the JGI for sequencing. While JGI processed, sequenced, and analyzed the soil samples, the JGI offered 7 seminars to the class throughout the project life cycle. During regular web conferences, various JGI subject matter experts walked the class through the workflow and all the process steps of sequencing and analyzing the DNA samples. The class was also able to receive training for the JGI’s Integrated Microbial Genomes & Microbiomes (IMG/M) portal for their sample analysis.
The class had to sift through over 55 billion genes and 16 trillion DNA bases of data, to figure out what to study and compare for the project. At the same time, in order to analyze and compare their soil, the class partnered with Texas A&M researchers to assist them with soil sample analysis for nitrogen and carbon concentrations along with water content.
The class had to work collaboratively and creatively to come up with a sense of what to focus on. With so much data and a lack of experience, this added a layer of complexity to the project for figuring out how to bring this project together. The class understood that much is unknown about the taxonomic diversity and methane metabolism within the Florida Everglades. They decided to look at the predominant taxa and the abundance of genes involved in environmentally significant metabolic pathways related to methane production, nitrogen fixation, and dissimilatory sulfite reduction.
Presenting the Results
The Boca Raton A-Level AICE Biology Class was invited to present the results of their efforts during the Energy, Earth and Environmental Education (E4) community-building workshop at the 2019 Association for Environmental Studies and Sciences (AESS) Conference in Orlando, Florida. The JGI was also invited to participate in the workshop and speak about this special partnership with Boca Raton Community High School.
On June 26, 2019, the students gave a team presentation on how they defined, prepared, and carried out their research and analysis for this project. They had learned many statistical tests for raw data and how to use some bioinformatics tools for analysis to help them focus on what to pull out from the enormous data set on the IMG portal. The class discovered Actinobacteria, Acidobacteria, and Proteobacteria to be the most common phyla of bacteria sequenced from the metagenomic soil samples from the four sites tested. They found Euryarchaeota to be the most common phylum of Archaea. Alpha/Beta diversity tests showed significant congruity between three sites. There was slight congruity between all the sites. Shotgun metagenomic analysis discovered the presence of integral biomarkers: mcrA, nifH, and dsrB. Lastly, for this project the students calculated the correlations between the integral biomarkers and abiotic factors for water content, nitrogen concentration, and organic carbon concentration.
From Field Studies to Publication
Together as a class, the students learned about how to write a scientific paper. They wrote, edited, and rewrote a microbiome announcement that has been submitted to the journal Environmental Microbiome with significant scientific observations for this Everglades area. Their paper is currently under peer review. Although the class is over and the students are preparing for their senior year, they will have to come back together as the paper goes through additional review and revisions. It was incredibly exciting and inspiring to see what these students accomplished in such a little amount of time and with minimal direction.
During the presentation, students were asked what they would do differently if they did the project again. Some mentioned that they might sample the west side because it was close to where there was agricultural run-off. They were wondering how that might compare with the east side, and if the run-off might have affected the results. Another idea was to study the different methane-producing bacteria and even how some of the bacteria could be competing with each other. One student thought that there could be an undiscovered gene that was sequenced in the project to breakdown methane and noted how awesome it would be to discover that gene from this untouched Everglade region.
“The JGI has gone over-and-above throughout this research project,” said Jonathan. “Students now have a deeper understanding of both the research process and metagenomics while legitimately contributing to the body of scientific knowledge that can be accessed by the rest of the community. These high school students now feel more empowered and equipped for their future research endeavors.”
The students were able to do professional-level research with hands-on experience for soil collection, processing, and DNA extraction. As a team, they learned and showed invaluable leadership, teamwork, and dedication. This was a first of its kind project, but it is a great model of how the JGI can partner and help train the next generation of scientists.