The game where one has to guess how many jelly beans or marbles can fill a jar should never be played with the cyanobacterium Prochlorococcus. By some estimates, in a single liter of water as many as 100 million cells of this tiny bacterium can be found. These important organisms serve as the base of the ocean food chain and are thought to be responsible for providing about 20% of the oxygen produced by the planet each year.
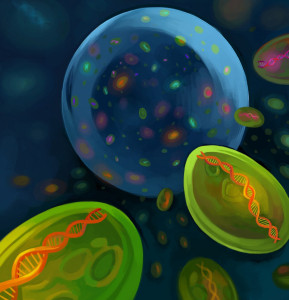
MIT scientists discovered an amazing amount of diversity among a population of the marine microbe Prochlorococcus found in a few drops of water. Each subpopulation is characterized by a shared genomic “backbone” and a few flexible genes. The figurative backbone is depicted in this artist’s rendering of the bacteria and their diversity in a single drop of seawater. (Carly Sanker, MIT)
Though it is considered a single species, this unicellular organism can be classified into several distinct major ecotypes – characterized by variable factors such as seasonal, depth, and geographic patterns. However, even within these ecotypes, Prochlorococcus cells still display a wide range of genomic diversity. To learn more about the cyanobacterial populations at this level, MIT marine microbiologist Sallie Chisholm, a longtime collaborator of the U.S. Department of Energy Joint Genome Institute (DOE JGI), led a team that applied single-cell genomics to these cyanobacteria collected from the same environment at three separate times of the year.
As reported in the April 25, 2014 issue of Science, Chisholm, her post-doctoral fellow Nadav Kashtan, and their collaborators at the DOE JGI sequenced and assembled Prochlorococcus genomes from single cells collected at the Bermuda-Atlantic Time-series Study site in the northwestern Sargasso Sea between November 2008 and April 2009. They found cell clusters within known ecotypes that indicated that the relative abundance of cyanobacterial subpopulations shifted along with the seasons, likely in response to environmental changes. They then analyzed the cells on a finer scale, sequencing partial genomes from select clusters.
The team wondered if the Prochlorococcus subpopulations within the clusters were genomically distinct. The answer turned out to be yes, and each of these so-called “genomic backbones” is comprised of highly conserved core gene alleles, or alternative forms, and a smaller distinct set of flexible genes. “This covariation between the core alleles and flexible gene content, and its fine scale resolution, represents a new dimension of microdiversity within wild Prochlorococcus populations,” the team reported in their study. “Similar patterns have been identified in cultured isolates and metagenomic assemblies within coexisting members of a few other microbial species with very different ecologies, suggesting that differentiated genomic backbones may be a feature of diverse types of microbial populations.”
The vast diversity of these abundant Prochlorococcus subpopulations was validated with help from the DOE JGI’s Rex Malmstrom. “My contribution was to provide the essential control data,” he explained. “We could do this based on our extensive R&D work on single cell genomics.”
Malmstrom said that the Chisholm lab found substantial sequence variation from cell to cell in wild Prochlorococcus populations, but needed controls to understand what fraction of the differences among individual Prochlorococcus cells was due to the genome amplification, sequencing, and assembly process. This could be determined from the analysis of many known-to-be-identical individual cells using the same sample analysis pipeline.
“This is a difficult and time consuming thing to do, but it’s the sort of thing we do regularly at the JGI when we test out changes to the single cell procedure,” he said. “So I looked through our R&D data on E. coli, our test organism of choice, and found a set of data that underwent similar treatment and had a similar sequence quality to the data used for the wild Prochlorococcus. We estimated the differences among the identical E. coli to be 100-fold lower than the differences among wild Prochlorococcus using our own assembly and analysis procedures. For good measure, the Chisholm lab also ran the raw E. coli sequence data through the same assembly and statistical analysis procedure they used for Prochlorococcus and found differences among the identical E. coli to be 100-fold lower than the differences among wild Prochlorococcus. This established that the magnitude of the differences they saw among the wild Prochlorococcus cells was by far the strongest signal in the data and technological artifacts were minimal.”
The team noted that if each of the cyanobacterial backbone subpopulations were counted as distinct species, then Prochlorococcus consists of thousands of species. Further, they added, each of these groups likely helps maintain the “dynamic stability of the Prochlorococcus ‘collective’ in the global oceans.” Such a large set of coexisting subpopulations with distinct genomic backbones, the team concluded, may be characteristic feature of free-living bacterial species with huge populations in highly mixed habitats.
“It was heartening to work with JGI on part of this project because that is where the Prochlorococcus genomics story began about 15 years ago,” said Chisholm. “They sequenced the genomes of two of our strains that had been isolated from different oceans, giving us our first peek at the diversity within this group. Likewise, JGI’s current collection of a number of Prochlorococcus genomes has been an invaluable resource in our more recent work. These ‘reference genomes’ are key to unlocking the mysteries of the patterns we see in the genomes of wild populations.”