Genome sequences provide clues to the evolution of sensory perception
Fungi can sense environmental signals and react accordingly, changing their development, direction of growth, and metabolism. Sensory perception lies at the heart of adaptation to changing conditions, and helps fungi to improve growth and recycle organic waste, and to know when and how to infect a plant or animal host. New results based on characterizing and then conducting a comparative analysis of two genome sequences published online May 26, 2016 in the journal Current Biology shed light on the evolution of sensory perception in fungi.
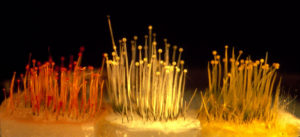
Phycomyces fungi, showing sporangiophores (fruiting bodies) in the wild type and color mutants. (Photo by Tamotsu Ootaki.)
For the study, researchers sequenced and annotated Mucoromycotina genomes, specifically those of Phycomyces blakesleeanus and its relative Mucor circinelloides. The work was led by researchers at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, in collaboration with scientists from 31 research centers and universities from 13 countries, and coordinated by scientists from the University of Seville in Spain. Capturing the genomic variation of fungi allows researchers to build a foundation for translating their genomic potential into practical applications. For example, understanding the mechanisms by which these environmental cues are sensed could provide insights on how some fungi can change their growth patterns to act as pathogens rather than benign organisms.
Understanding fungal metabolism
“Very little is known about basal fungi such as Mucoromycotina and genomics may be the most efficient way to understand their metabolism,” said DOE JGI Fungal Genomes program head Igor Grigoriev of this project, which was done as part of a 2006 Community Science Program project. “Many members of this phyla show very high sensitivity to environmental signals, which when understood could be used for natural control of some of these metabolic processes. The high capacity for accumulating lipids in Mucor circinelloides, for example, may have biofuels applications.”
The team found that both species had undergone whole genome duplication, a rare occurrence in fungi. The genome duplication led to an expansion of gene families and the development of specialized genes, providing new proteins that have enabled these fungi to refine the way they perceive signals from the environment to regulate their growth and development.
Evolution of sensory perception

The fruiting bodies (sporangiophores) of Phycomyces blakesleeanus grow out of the mycelium and reach several cm in length. The speed and direction of growth is controlled by signals from the environment including light, gravity, touch, wind, and the presence of nearby objects. The ball at the top of each fruiting body is the sporangium with spores. (Photo by Maria del Mar Gil Sanchez)
Biologists have been fascinated for well over a century by the finely-tuned sensory responses of the sporangiophores of P. blakesleeanus. The sporangiophore is a long, rapidly growing giant cell with many nuclei that responds to light, gravity, touch, and even the mere presence of nearby objects. These “spore carriers” are special aerial hyphae that grow into the air for several centimeters and disperse the spores formed at their tip. P blakesleeanus approximate the sensitivity of the human eye to both dim light of stars at night, and the luster of mid-day in bright sunlight.
“Phycomyces blakesleeanus is best known because it was studied by Nobel laureate Max Delbrück, who recognized its potential as a model organism for the study of sensory perception,” noted University of Seville researcher Luis Corrochano, the study’s first author and coordinator of the project. “Delbrück was attracted by the elegant simplicity of the behavior of the fruiting body from P. blakesleeanus that can react to light and other signals from the environment, including gravity, wind and adjacent objects by changing its direction of growth.” Fittingly, the researchers dedicated this study to Delbrück’s memory.
Mucor circinelloides, a related fungus, has similar environmental responses, and sometimes also acts as a human pathogen. The genome sequences of Phycomyces and Mucor provide evidence that an ancient genome duplication yielded new genes to expand signal transduction pathways and improve the mechanisms for sensing light and other signals in this group of fungi. For example, the team found that exposure to light produced massive changes in gene regulation in different fungal tissues and hypothesized that the refined response to light is a consequence of gene duplication and specialization after whole genome duplication. These results advance understanding the role of genome dynamics in the evolution of sensory perception which, in turn, could provide leads to genes and pathways useful for understanding fungal adaptation and for accelerating the breakdown of biomass for bioenergy.