A new system called ScanDrop could revolutionize how we identify pathogens in drinking water.
The Science:
Researchers from Lawrence Berkeley National Laboratory (the Joint BioEnergy Institute and the DOE Joint Genome Institute) and Northeastern University and the Massachusetts General Hospital/Harvard Medical School) have developed a portable, network-enabled system for testing drinking water contamination. The system, called ScanDrop, developed by Tania Konry’s group (NEU/MGH/HMS), uses droplet based microfluidics and bead based assay technologies with integrated portable optics to detect bacteria in water. This ScanDrop system was combined with automated microscope control software (PR-PR) and cloud-based networking, developed by Nathan Hillson’s (LBNL/JBEI/JGI) group, to scan water samples for pathogens and transmit the data remotely.
The Impact:
Testing drinking water supplies for disease-causing pathogens requires several days to test a water sample in a laboratory. This study serves as a proof-of-concept for a method of testing for pathogens in drinking water that is faster than current options and cheap enough that it could be deployed in many poor countries. The system could also potentially be set up to test for several pathogens at once or to test for other kinds of contaminants, including environmental sampling for bacteria that impact the global carbon cycle or are used for bioremediation.
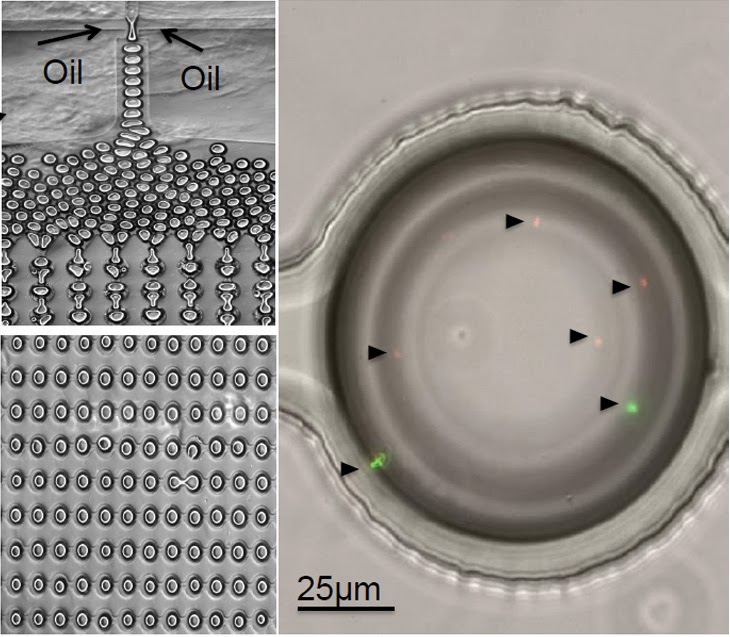
On the left, a graphic demonstrating how droplets of oil move through ScanDrop’s microfluidic chip. On the right, the arrows indicate single bacteria cells inside a droplet of oil. (Image courtesy of N. Hillson)
Summary
Contaminated water is a major source of infectious illness. Current systems used to test for disease-causing pathogens in drinking water require taking a water sample to a laboratory for testing, which usually requires a minimum of two to four days. More advanced systems exist, but they are either too expensive or not portable enough to be widely used, especially in poor countries most likely to have pathogen-contaminated drinking water.
In a paper published January 22, 2014 in the journal, PLOS ONE, the team introduces a new alternative, ScanDrop. It uses several recent advances to assemble a system for flagging the bacteria, optically scanning for them and transmitting the information to interested parties, all in a matter of hours. The bacteria are captured from the drinking water sample using magnetic beads. Then, they are passed through a microfluidic chip a few centimeters wide that uses a few hundred picoliters of oil to coat the bacteria, much like bugs trapped in amber. A portable, remotely controlled microscope is used to optically scan the output from the chip and the results are relayed via “cloud” network to a control center, and from there to public health professionals. This process takes about eight hours, a fraction of the time required for current methods for testing for pathogens in potable water.
Several ScanDrop test systems could be set up at various points in a drinking water system to track water quality in a large system. Moreover, it has the potential to be adapted to test for different pathogens. In the future, a network of ScanDrop testers could be set up to continuously monitor a drinking water system and rapidly identify contamination breaches.
Contact
Nathan Hillson
DOE JGI
njhillson@lbl.gov
Publication
Golberg A, Linshiz G, Kravets I, Stawski N, Hillson NJ, et al. (2014) Cloud-Enabled Microscopy and Droplet Microfluidic Platform for Specific Detection of Escherichia coli in Water. PLoS ONE 9(1): e86341.
doi:10.1371/journal.pone.0086341
Funding
Department of Energy, Office of Science
Shriners Foundation
Singapore National Research Foundation
Campus for Research Excellence and Technological Enterprise
Related Links
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0086341