Proposals aim to sequence and annotate genomes from Antarctica to Africa to global oceans.

Laura Selbman of Italy’s University of Tuscia proposes to sequence nearly 100 species of black fungi in a 3-year effort called STRES (Shed light in The daRk lineagES of the Fungal Tree Of Life (FTOL)). Black fungi are ubiquitous in many environments – the collage shows examples of habitats colonized by these fungi – and extremely tolerant of stresses ranging from UV radiation to heat to toxic metals and organic pollutants.(Collage courtesy of L. Selbmann)
Through the Community Science Program of the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science user facility, 24 large-scale proposals have been accepted from 70 full submissions based on 92 letters of intent. Additionally, 40 percent of the proposals were submitted by researchers who had not been a primary investigator on any proposals previously accepted through JGI’s calls.
Nearly 200 Terabases of data are expected to be generated for the accepted proposals, said Susannah Tringe, JGI User Programs Deputy. “These new CSP projects accelerate our efforts to characterize poorly-studied algal and viral genomes, and integrate with key DOE Office of Biological and Environmental Research (BER) initiatives in bioenergy crop improvement and terrestrial carbon transformations.”
The JGI’s large-scale call included algal genomics as a focus area, and one of the accepted proposals came from Chris Bowler of France’s Institut de Biologie. His team proposed to sequence Chaetoceros microalgae, or diatoms. Through the Tara Oceans Expeditions that took place between 2009 and 2013, the Chaetoceros genus has been identified as the most abundant in the world’s oceans, found in a wide range of morphologies and ecologies. Diatoms, Bowler’s proposal noted, are believed responsible for a fifth of the planet’s photosynthesis activities. Aside from being a key component of the marine food web base, they also feature in many nutrient cycles. Thus, having reference Chaetoceros genomes would provide more information on these ecologically relevant species.
Another focus area was inter-organismal interactions, and two fungal proposals focus on symbiotic relationships. Jessie Uehling of the University of California, Berkeley is interested in better characterizing the role endosymbiotic bacteria play on mating habits of Mucoromycota fungi, and their effects on these oil-producing species. Many fungi have beneficial symbiotic relationships with plants and bacteria; in many instances, fungal physiology is impacted by these bacteria during mating processes. Kathryn Bushley of the University of Minnesota is interested in learning more about the fungal endophytes of soybean and how they deter two significant pathogens, the soybean cyst nematode and the soybean sudden death pathogen.
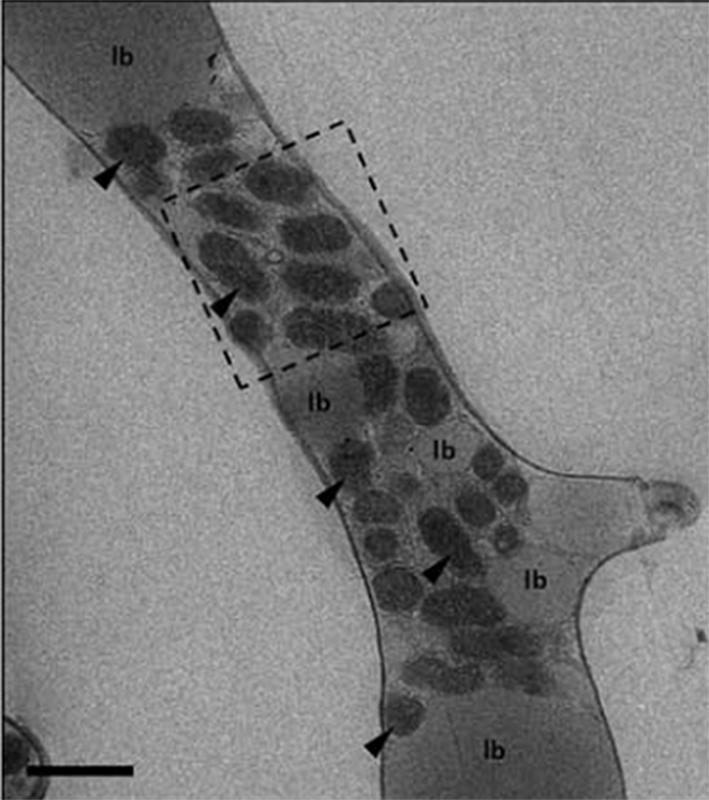
Related to Uehling’s proposal: Transmission electron microscopy of Mycoavidus cysteinexigens (arrows) aggregated around lipid bodies (lb) inside hyphae of Mortierella elongata isolate NVP64. Scale bar 1 um. (Alessandro Desiro)
In the genes-to-function focus area, Daniel Kliebenstein from the University of California, Davis proposes to develop high quality reference genomes for 10 individuals from each of the following five species: Camelina sativa, Thlaspi arvense (pennycress), Crambe hispanica, Brassica oleracea, and Streptanthus barbigeri (bearded jewelflower). Part of efforts to develop pangenomes for these species, this approach would also allow researchers to learn more about what is and is not modifiable in crops, which can help optimize crop improvements.
Some of the proposals were associated with more than one focus area of the proposal call. Richard Dick of The Ohio State University aims to study plant drought resilience in the Sahel, an arid region of West Africa. The staple crop there is millet, grown without fertilizer and under erratic rainfall conditions. To help mitigate drought, famers are growing millet with an indigenous shrub and these intercropping efforts appear to be redistributing water availability. By sequencing the millet rhizosphere and endosphere communities both with and without the shrub’s presence, researchers hope to learn more about the mechanisms that allow the millet to thrive in drought conditions. The proposal targets the CSP focus area in plant microbiomes, as well as those in inter-organismal interactions and terrestrial elemental cycling.
George diCenzo of Canada’s Queens University aims a multi-omics approach toward generating “with unprecedented precision” the gene functional networks and regulatory networks of a single strain of Sinorhizobium meliloti. S. meliloti is one of the rhizobia, a group of nitrogen-fixing bacteria with symbiotic relationships in plant roots of legume crops such as soybeans and peas. Their presence reduces the need to add fertilizers to the soil.
diCenzo’s proposal is also notable as he had an accepted new investigator proposal last year, and he noted that the experience proved beneficial for the large-scale call. “I am convinced that the experience writing the New Investigator proposal, and receiving feedback from reviewers, was invaluable in preparing the large-scale application. It provided practice in writing for the JGI and describing my general research area and its relevance to the JGI, and the feedback from reviewers allowed me to be better prepared and to avoid making the same mistakes,” he said. “Although in my specific case the projects are different for the two proposals, I can see the New Investigator program being a valuable “stepping-stone” to a large-scale proposal by allowing researchers to test a project on a smaller scale as a proof-of-concept or by allowing the generation of preliminary data for a larger study.”
The accepted proposals start October 1, 2019. See the full list of accepted proposals at https://jgi.doe.gov/our-projects/csp-plans/approved-proposals-fy20/.