
From left to right, several of the authors of the ProDeGe paper published in The ISME Journal: Nikos Kyrpides, Scott Clingenpeel, Kristin Tennessen, Tanja Woyke, Amrita Pati, and Evan Andersen. Tennessen will talk about ProDeGe at the September 2015 Microbial Genomics & Metagenomics Workshop at the DOE JGI.
Single cell genomics and metagenomics are pioneering techniques that have helped researchers assess environmental microbial community structure and function. As projects applying these techniques scale up, however, researchers are hindered by the lack of a high-throughput process to review assembled genome sequences. Currently, sequence decontamination of the microbial genomes being uploaded to public databases is a manual and time-consuming process that requires information about the contaminant sequences in order to remove them.
To help resolve this obstacle, a team from the Prokaryotic Super Program at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, has developed the first computational protocol for quick and automated removal of contaminant sequences from draft genomes. They describe the tool called ProDeGe (Protocol for Decontamination of Genomes) in a study published online June 9, 2015 in The ISME Journal. The paper has made The ISME Journal‘s Top Ten chart of most downloaded articles since its release.
Though the team says ProDeGe works on any type of genome sequence, for the study, it was benchmarked using 182 manually screened single amplified genomes (SAGs) from two publicly available datasets, one of them the Microbial Dark Matter project and the other using Arabidopsis endophyte data enabled by collaborators at the University of North Carolina, who are coauthoring this paper.
Speedy Sequence Decontamination
The tool classifies sequences as either “clean,” or “contaminant,” and runs, the team reported, at a rate of 0.30 CPU core hours per megabase of sequence. “It takes an expert approximately six hours to manually decontaminate 1 megabase of sequence,” noted the study’s first author Kristin Tennessen, “so using ProDeGe results in a speedup of about 20 times.” If the manual user is inexperienced, she added, the increase in the decontamination rate can be even greater.
Nikos Kyrpides, head of the Prokaryote Super Program at the DOE JGI, said that the emergence of software solutions–such as ProDeGe–to daunting computational challenges is consistent with one of the three “pillars” of the Institute’s 10-Year Strategic Vision. “With an emphasis on Biological Data Interpretation,” he added, “the DOE JGI has played a leadership role in developing, standardizing and providing access for users to high-quality genome assemblies, annotations and other computational genomics tools.”
ProDeGe is pre-calibrated to remove at least 84 percent of contaminant sequence, and the team found it performed best when it could compare the test sequence against homologs in the database that corresponded at the Class level or deeper. If the sequences belong to novel organisms, the team reported, ProDeGe removes contaminants solely by checking the sequence composition.
Sequence Decontamination Tool for Quality Control
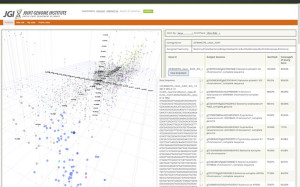
The ProDeGe web service at prodege.jgi-psf.org allows users to upload their samples for processing. Users view their contigs in a k-mer PCA plot and can drill down for further metadata such as assigned taxonomy, sequence, gene hits, and clean/contaminant classification.
“Given the enormous volume of environmental sequence information generated each year and the increasing popularity of single-cell genomic sequencing,” said Steven Hallam, a longtime DOE JGI collaborator at the University of British Columbia and a ProDeGe user. “ProDeGe fills a critical gap in QA/QC workflows that actually scales effectively between individual users and platform services.”
The research team added that, “ProDeGe is the first step towards establishing a standard for quality control of genomes from both cultured and uncultured microorganisms. It is valuable for preventing the dissemination of contaminated sequence data into public databases, avoiding resulting misleading analyses. The fully automated nature of the pipeline relieves scientists of hours of manual screening, producing reliably clean datasets and enabling the high-throughput screening of datasets for the first time. ProDeGe therefore represents a critical component in our toolkit, during an era of next-generation DNA sequencing and cultivation-independent microbial genomics.”
Speaking as one who has used the ProDeGe tool, Ramunas Stepanaukas, director of the Bigelow Laboratory Single Cell Genomics Center, and a DOE JGI collaborator added that, “Single cell genomics and metagenomics have become major sources of information about the biology of the uncultured microorganisms, which are the predominant component of most ecosytems on our planet. The risk of DNA contamination is a significant challenge to both single cell genomic sequencing and metagenome assemblies. The prevention, detection and removal of contaminants from single cell genomics and metagenomics data is of key importance for understanding the ecosytems on our planet. Novel laboratory and computational tools, such as ProDeGe, will be critical to ensure high standards of data quality in these emerging research fields.”
ProDeGe’s web interface for uploading and analyzing datasets can be found at http://prodege.jgi-psf.org. Standalone software for ProDeGe can be downloaded from http://prodege.jgi-psf.org/downloads/src can be run on a system with Perl, R, and NCBI Blast. Tennessen will be presenting on ProDeGe during the next Microbial Genomics & Metagenomics (MGM) Workshop, taking place September 14-18, 2015 at the DOE JGI.