The U.S. Department of Energy Joint Genome Institute (DOE JGI) and the Environmental Molecular Sciences Laboratory (EMSL) have accepted 10 projects submitted during the 2017 call for proposals for their joint “Facilities Integrating Collaborations for User Science” (FICUS) initiative.
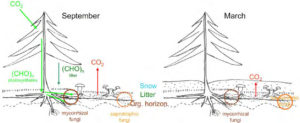
This diagram from Petr Baldrian’s proposal shows the seasonal differences in the carbon cycle processes in the temperate and boreal coniferous forests. During vegetation seasons, depicted by September on the left, photosynthesis products are allocated to soil via tree roots. When photosynthesis stops in winter, depicted by March on the right, decomposition is the most important carbon cycle process. (Image courtesy of Petr Baldrian)
These new research projects all involve collaboration between two user facilities that are stewarded by the DOE Office of Biological and Environmental Research and integrate the expertise and capabilities to address pressing questions in support of DOE’s energy, environment and basic research missions. The FICUS initiative represents a unique opportunity for scientists to harness the combined power of genomics and molecular characterization in a single proposed research project. The researchers, whose projects were selected by outside reviewers, will access the capabilities of both user facilities to generate datasets unique to these two providers of critical infrastructure – deriving benefits beyond what could be generated by either facility by itself.
The accepted proposals will begin on October 1, 2016 and fall under the following focused topic areas: Plant-Microbe Interactions, Biofuels and Bioproducts, and Biogeochemistry of Select Inorganics.
Several projects focus on the response of soil microbial communities in various ecosystems:
- Jeffrey Blanchard of University of Massachusetts Amherst and his colleagues aim to determine which metabolic processes are active in soil communities in response to increased temperature, and which microbes are responsible for increased carbon dioxide flux through heavy water labeling of the active biota. How soil microbial communities respond to climate change could change the soil’s role from carbon sink to carbon source. Understanding the ecological function of soil communities in their natural environment is essential in ecosystem modeling.
- Virginia Rich of The Ohio State University is interested in the implications of the predicted thawing of the northern permafrost. Using a well-studied “model ecosystem” of permafrost thaw she and her colleagues intend to look at the pathways for carbon loss in thawing permafrost, and metabolic changes in peatland microbial communities that are regulating the carbon flux.
- Kelly Wrighton of The Ohio State University is interested in how warming temperatures are allowing more shrubs to grow in the Arctic, which results in an increase in condensed tannins and other molecules that could inhibit microbial degradation of carbon. To learn more about the impact of these molecules on the soil and ruminant microbial communities found in the regional ecosystem, she and her team aims to use Alaskan willows to recreate a model condensed tannins pathway that can be used to test relevant arctic microbial communities.

In this diagram from Virginia Rich’s proposal, the team aims to characterize extracts of the litter of the dominant plant species in each habitat. Permafrost thaw drives successional changes in the overlying plant communities, with dramatic shifts in dominant species. These in turn change the amount and composition of new carbon inputs to the soil column. (Image courtesy of Virginia Rich)
Two projects involving soil communities are focused on the interactions involving conifers and ecotmycorrhizal fungi:
- Petr Baldrian of the Academy of Sciences of the Czech Republic is studying soil and leaf litter microbes in coniferous forests to learn more about nitrogen availability on carbon cycle-related processes. Through metabolomics analyses, researchers hope to identify drivers of seasonal changes in microbial function — both inside and outside the tree root — as well as the interplay between tree roots and their symbiont fungi. Additionally, the fungal genomes that are being obtained through the DOE JGI’s 1000 Fungal Genomes Program are expected to provide ecological perspectives.
- Hui-Ling Liao of Duke University is studying the symbiotic interactions between conifers and ectomycorrhizal Suillus fungi found in the roots and rhizosphere soil. This team aims to better define the roles these species play in soil nutrient turnover.
Other projects focus on coastal marine environments:
- Tom Bianchi of University of Florida is interested in a phenomenon known as the “priming effect” that is well-studied in soils but poorly understood in the marine environment. Along the Florida coastline are mangroves, salt marshes and seagrasses that trap “blue carbon” or organic carbon derived from and stored in coastal and marine environments. Lab experiments show that in the presence of highly reactive substrates, dissolved organic carbon breaks down up to 75 times faster during the first 24 hours. One of the team’s questions is how the priming effect might influence blue carbon stores impacted by rising sea levels and the corresponding loss of wetlands.
- Rose Ann Cattolico of the University of Washington is interested in the alga Chrysochromulina tobin and its 10-member bacterial biome, 8 of which have already been sequenced. The team aims to understand the roles of this alga-bacterial consortium in processes including carbon fixation and nitrogen processing, as well as how changing ocean salinity impacts the metabolic interplay in this alga-bacterial consortium.
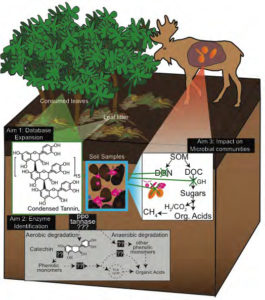
In this diagram from Kelly Wrighton’s proposal, willow leaves containing condensed tannins (CT) are eaten by moose or become part of the leaf litter, and impact the soil (pink) and rumen (orange) microbial processes. Data gathered from this study would shed light on additional enzymes, metabolites and pathways aside from CT pathways, shown in the gray box. (Image courtesy of Kelly Wrighton)
Two projects are focused on generating biofuels from biomass:
- Laura Bartley of the University of Oklahoma is interested in secondary cell wall development in grasses. While grass cell walls are among the most abundant potential biomass sources for biofuel production, the process of breaking down cell walls limits available biomass for conversion to sugars.
- Jeffrey Skerker of UC Berkeley wants to understand how biomass-derived carbon is converted into lipids and terpenoids in the yeast Rhodosporidium toruloides, and how carbon flux is regulated in this organism. Through a systems biology-level study, his team aims to develop R. toruloides into a novel metabolic engineering platform organism that can convert biomass-derived products into biofuels and bioproducts.
One project is focused on biogeochemistry:
- Scott Fendorf of Stanford University is interested in the persistence of contaminants in groundwater. Focusing on the Colorado River Basin, where legacy contamination has left floodplains with uranium traces, his team aims to learn about the metabolic constraints of organic matter decomposition in floodplain sediments, and how this regulation of the carbon cycle impacts metal cycling.
The full list of approved projects is available at http://jgi.doe.gov/our-projects/csp-plans/ficus-plans-fy-2017/.
***
EMSL, the Environmental Molecular Sciences Laboratory, is a DOE Office of Science User Facility. Located at Pacific Northwest National Laboratory in Richland, Wash., EMSL offers an open, collaborative environment for scientific discovery to researchers around the world. Its integrated computational and experimental resources enable researchers to realize important scientific insights and create new technologies. Follow EMSL on Facebook, LinkedIn and Twitter.