Brachypodium model system traces polyploid genome evolution.

Brachypodium distachyon, the model species for temperate cereals and biofuel crop grasses with a growing pan-genome of one hundred genomes, in Spain: Huesca, Ibieca, San Miguel de Foces. (Pilar Catalán)
Flowering plants abide by the concept, “the more the merrier,” with respect to their genomes. In their base state, they are diploids with two genome copies, one from each parent. Having three or more genome copies from additional parents or duplication, also known as “polyploidy,” is common among flowering plants; at least once during their evolution, the genomes of flowering plants multiply. Over time, plants lose many genes after such events, returning their genomes to a diploid state while retaining multiple copies of some genes.
This polyploidization process is ongoing and many plants, including agricultural staples and candidate bioenergy feedstock crops, are recent polyploids. Crop breeders have harnessed polyploidy to increase fruit and flower size, and confer stress tolerance traits. Given the importance of polyploidy in plants, researchers have sought to learn the origins, evolution and development of plant polyploids. A multi-institutional team led by researchers at Spain’s Universidad de Zaragoza and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), relied on a model grass system for answers. Their work recently appeared in Nature Communications.
The team studied Brachypodium hybridum, a polyploid resulting from the combination of the genomes from the diploids B. stacei and B. distachyon, a relative of the candidate bioenergy crop switchgrass and a JGI Flagship Plant. All three Brachypodium grasses have small genomes, short life cycles and can be easily transformed, traits that make them easy to work with bioinformatically and in the laboratory. “We are fortunate because the progenitor species of Brachypodium hybridum are still extant (whereas the diploid progenitors of other polyploid plants have gone extinct or are unknown),” noted Pilar Catalan of the Universidad de Zaragoza, one of the paper’s senior authors. “It offers us the possibility to explore genomically the whole complex of progenitors and descendant species and, consequently, the evolution of polyploidy.”
Evaluating Polyploid Genomes in a Pan-genomic Context
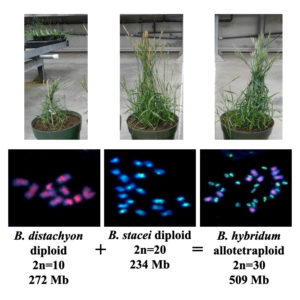
Plant images from the Vogel lab. Cytogenetic images from Hasterok et al. 2004, 2006 (Courtesy of John Vogel)
For the study, partly enabled by the JGI’s Community Science Program, the team generated reference genomes for B. hybridum and B. stacei, as well as multiple lower quality genome assemblies for all three grasses. With these datasets, researchers made a pan-genome containing four B. hybridum genomes and many B. distachyon genomes. This builds upon a previously made B. distachyon pan-genome.
A pan-genome is the nonredundant set of genes found in a species, but in this case, the team included two species in the pan-genome. Typically, pan-genomes are much larger than any individual genome highlighting the fact that the genetic information in a single reference genome is insufficient. “We really have to evaluate polyploid genomes in a pan-genomic context so we don’t overestimate the evolution that happened after polyploidization,” said John Vogel, head of the Plant Functional Genomics program at the JGI and a senior author on the paper. “The assumption that all the genes in a diploid reference genome were present in the actual diploid progenitor of the polyploid reference line is simply not true based on previous pan-genome studies. Thus, we must compare multiple polyploid genomes to the pan-genome to better estimate post-polyploidization evolution.”
One Polyploid Species, Two Subgenomes
Among the team’s findings is that the polyploid B. hybridum formed more than once, and these events took place a million years apart: the individual with a maternal B. distachyon parent (D plastotype) some 1.4 million years ago (Mya), while the S plastotype was a more recent event (0.14 Mya) with a maternal B. stacei parent. The analysis also revealed that these two B. hybridum types don’t interbreed. “Preliminary artificial crossing experiments between ancestral and recent B. hybridum accessions produced infertile F1 descendants (first offspring of the parents), posing the question if they correspond to the same recurrent speciation outcome or to different species,” added Catalan.
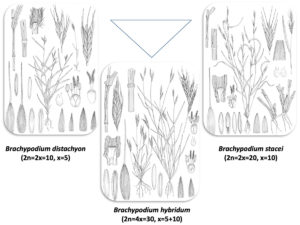
The Brachypodium distachyon-B. stacei-B. hybridum polyploid model complex.
(Illustrations by Juan Luis Castillo)
Overall, Vogel noted, the results suggest that polyploid evolution is occurring slower than had been expected. “Although every case is probably going to be different, at least for B. hybridum it’s happening a lot slower than I had anticipated with only minor gene loss even after a million years.”
Reference genomes for B. distachyon, B. stacei and B. hybridum are all available on the JGI’s plant portal Phytozome v13.
Researchers from the following institutions were also involved in this work: Estación Experimental de Aula Dei (EEAD-CSIC) (Spain); Fundación ARAID (Spain); University of California, Berkeley; Gregor Mendel Institute of Molecular Plant Biology (Austria);
Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben (Germany); Université d’Evry Val d’Essonne (UEVE) (France); Zhejiang University (China); Universidad de Zaragoza-Escuela Politécnica Superior de Huesca (Spain); Universidad Central de Venezuela (Venezuela); University of Silesia (Poland); Aberystwyth University (UK); HudsonAlpha Institute for Biotechnology; University of Massachusetts Amherst; and, Tomsk State University (Russia).
Publication: Gordon SP et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat Commun. 2020 July 29. doi: 10.1038/s41467-020-17302-5
Byline: Massie S. Ballon