A team has optimized a crucial part of CRISPR-Cas9 technology to enable improvements in microbial oil production.
The Science
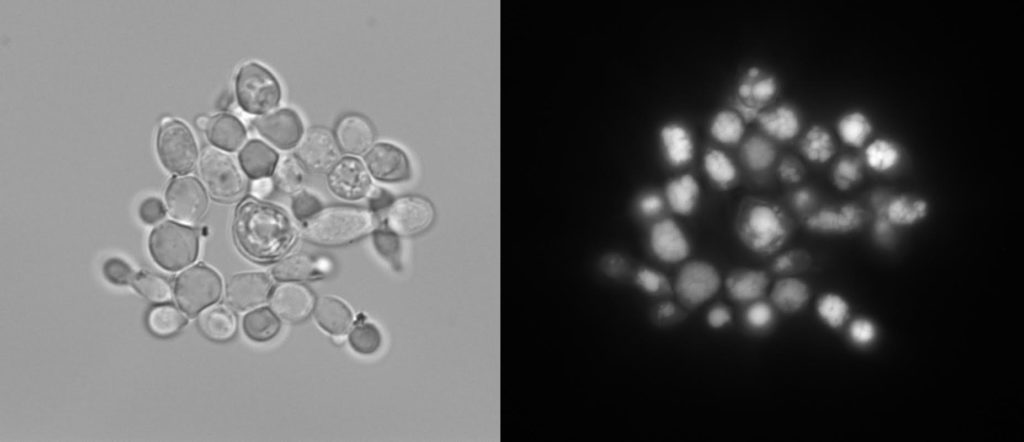
The lipid producing yeast, Yarrowia lipolytica, examined with light microscopy (left) and fluorescence microscopy (right), after being stained with Nile Red to visualize the lipid droplets inside (shown here in white). (Courtesy of Hal Alper)
CRISPR-Cas9 is a powerful, high-throughput gene-editing tool that can help scientists engineer organisms for bioenergy applications. Cas9 needs guide RNA to lead it to the correct sequence to snip — but not all guides are effective. To ensure the gene editing tool will make the right cut, researchers created a set of guide RNAs that were effective against 94 percent of the genes in a lipid-prolific yeast.
The Impact
The team used this set of guide RNAs on Yarrowia lipolytica to identify essential genes and genes that lead to increased lipid production. These insights into Y. lipolytica biology and lipid metabolism furthers the U.S. Department of Energy’s goal of producing sustainable fuels from alternative sources. Moreover, the team says the approach they used to validate their set of guide RNAs could be applied to other non-conventional organisms.
Summary
According to legend, Abraham Lincoln once said, “Give me six hours to chop down a tree and I will spend the first four sharpening the axe.” In this study, published in Metabolic Engineering, a team of researchers likewise honed their tool before felling their research problem: at a genetic level, how does Y. lipolytica work?

Cory Schwartz, then graduate student at the University of California-Riverside, led the work to develop trusted single guide RNAs for CRISPR-Cas9 in Y. lipolytica. (Cory Schwartz)
The project was a multi-institutional effort led by then-graduate student Cory Schwartz and his advisor Ian Wheeldon, a chemical engineer at the University of California, Riverside. The team also included researchers at the U.S. Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility.
Y. lipolytica is a tantalizing microbial factory, capable of turning abundant biomass carbohydrates — glucose and xylose — into fatty acids that could be used for renewable fuel. In fact, lipids can make up more than 90 percent of the microbe’s dry weight.
In order to better harness these gobs of energy, a team of researchers sought to disrupt every gene in the genome with CRISPR-Cas9 in order to interrogate each one’s function. However, the team first needed to fix a defect in the approach: CRISPR-Cas9 relies on RNA with a short guiding sequence (20 bp) that directs the endonuclease where to cut, but these “single guide” RNAs (sgRNAs) can be faulty. Without a good sgRNA, Cas9 is ineffective and leaves the target gene intact: a false negative if the gene is actually essential.
Through the JGI’s Community Science Program, the scientists thus set out to identify which sgRNA sequences were reliable. In an elegant, quantitative screen, the team designed a pool of sgRNA sequences: six for every gene in the genome. The JGI DNA Synthesis Science team synthesized these sgRNAs, cloning them alongside CRISPR-Cas9 into an expression vector to create an sgRNA plasmid library.
The team was then able to screen for the most effective guides. To do this, they tested each guide in a Y. lipolytica strain lacking a necessary enzyme for DNA repair. Whereas an effective sgRNA would lead Cas9 to make a lethal double-strand DNA break, a dysfunctional sgRNA would be harmless. The team was thus able to measure sgRNA effectiveness from its lethality and choose only the most reliable sgRNAs for their final set.
Tool in hand, the team was ready to test what genes were essential to Y. lipolytica. They identified 1,377, or 17.5 percent, of Y. lipolytica’s genes to be required for growth. By contrast, an initial screen using an unvetted library (with both active and inactive sgRNAs) appeared to be rife with false negatives: that time, the team only found 359 genes to be essential.
The team also tested for genes that led Y. lipolytica to overproduce lipids — an important phenotype for biofuel production. They identified four genes, including one with no prior known function.
The project’s success demonstrates how to sharpen the CRISPR-Cas9 tool for a non-conventional eukaryotic host — an approach, the team says, that could be applied to other eukaryotes.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Ian Wheeldon, Ph.D.
University of California, Riverside
[email protected]
Funding:
This work was supported by DOE Joint Genome Institute grant CSP-503076 (IW, HA, and MB) and NSF 1706545 (IW). HA and JW were supported by the Office of Naval Research under grant N00014-15-1-2785. JW acknowledges additional support from the National Science Foundation Graduate Research Fellowship Program (DGE-1110007). SA, AB, and MB were supported by NSF 1706134. SL and WP were supported by NSF 1526742. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231.
Publication:
- Schwartz C et al. Validating genome-wide CRISPR-Cas9 function improves screening in the oleaginous yeast Yarrowia lipolytica. Metabolic Engineering. 2019 September. doi:1016/j.ymben.2019.06.007 [Epub ahead of print June 16, 2019]
Related Links:
- JGI Community Science Program
- JGI DNA Synthesis Science Program
- JGI DNA Synthesis Science Call
- CSP 2016 Proposal: CRISPR-enabled genome-wide screening of the oleaginous yeast Yarrowia lipolytica
- Ian Wheeldon at the 2018 JGI Genomics of Energy & Environment Meeting: http://bit.ly/JGI2018Wheeldon
By: Alison F. Takemura