Termites efficiently transform lignocellulose and humus into valuable sugars, fuels (hydrogen, methane), and other intermediates of interest for biotechnologists, by exploiting the metabolic capabilities of the diverse microbial symbionts inhabiting their hindguts. In the five evolutionary “lower” termite families, the hindgut microbial community is dominated by a unique assemblage of flagellate protozoa, which are important players in cellulose and hemicellulose hydrolysis and fermentation. The larger gut flagellates of lower termites harbor intracellular symbionts that represent a novel bacterial lineage at the phylum (division) level, the Endomicrobia. “Candidatus Endomicrobium trichonymphae” (CET) is the single most abundant and ribosome-active bacterial species present in the microbial species-rich hindgut of these insects. However, the biology of Endomicrobia, especially their function in the tripartite endosymbiosis between bacteria, protozoa, and termites, is completely unknown. Thus, the CET genome sequence may hold the key to unravel its true metabolic potential. In addition, the case study of Endomicrobia could provide important insights into the evolution of a bacterial endocytobiosis into cell organelles.
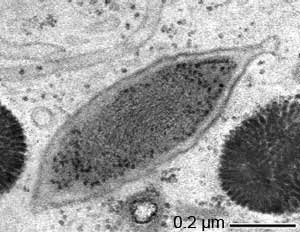
“Candidatus Endomicrobium trichonymphae”
Most strikingly, members of the Endomicrobia, like their cellulose-decomposing flagellate hosts, are entirely absent from the microbial community in the hindgut of all evolutionary “higher” termites. A separate CSP project (2005) examining the hindgut microbial community of a higher, wood-feeding termite (Nasutitermes sp.) from Costa Rica, identified Fibrobacteres as abundant and important members of the hindgut community. This project will allow the roles and functions of Endomicrobia and Fibrobacteres in the respective communities to be compared and contrasted, because they may hold a key to understanding the efficiency of lignocellulose conversion that occurs in these complex microbial bioreactors.
Our collaborators (at the MPI) recently isolated strain Pei191, representing a different class of probably nonendosymbiotic Endomicrobia so far obtained only as 16S rDNA clones from various environments. This sequencing project offers the unique opportunity to compare the genomes of a free-living and an obligately intracellular member of this phylum. Differences in metabolic potential betwee strain Pei191 and CET will provide insights into the evolutionary events leading to a symbiotic lifestyle.
Principal Investigators: Andreas Brune and Michael W. Friedrich (Max Planck Institute for Terrestrial Microbiology), Jared R. Leadbetter (Cal. Tech.), and Phillip Hugenholtz (JGI)