Analyzing the Role of DNA Methylation in a Bioremediation Bacterium
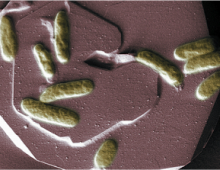
Characterizing functional roles in a microbe with bioremediation applications a first step toward similar studies for other prokaryotes The Science Researchers studied the role of DNA methylation on gene expression and other processes in the heavy-metal reducing bacterium Shewanella oneidensis MR-1 with the help of next-generation Single-Molecule Real Time (SMRT) sequencers from Pacific Biosciences. The… [Read More]