New high-throughput and cost-effective approaches provide a better understanding of microbiome functioning
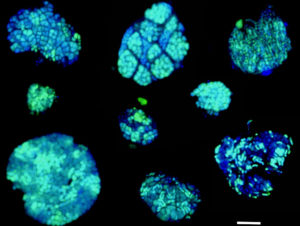
While typically thought of as solitary life forms, microbes frequently aggregate to form dense cell clusters. This often serves to achieve a common goal that cannot be reached by individual cells acting alone. The depicted cell consortia are composed of archaea and bacteria that, by combining their individual metabolic powers, together achieve oxidation of the greenhouse gas methane in the absence of oxygen. For the first time, these consortia have now been analyzed for their protein synthesis activity under a variety of conditions. In this image, nucleic acids within individual cells are stained in blue. Green color indicates that new proteins have been made. The scale bar equals 10 micrometer, or 1/8th the width of a human hair. (Credit: Roland Hatzenpichler)
Many uncultured microbes play unknown roles in regulating Earth’s biogeochemical processes; everything from regulating plant health to driving nutrient cycles in both terrestrial and marine environments, processes that can impact global climate. While researchers are harnessing multiple approaches to identify these microbes, referred to as “microbial dark matter,” and determine what they’re doing, most techniques don’t allow them to do both at once.
In a study published online June 28, 2016 in the Proceedings of the National Academy of Sciences (PNAS), researchers at the California Institute of Technology (Caltech) and the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, utilized a recently refined technique to identify both individual active cells, and single clusters of active bacteria and archaea within microbial communities. Understanding the true scope of the planet’s microbial diversity is of interest to the DOE in order to learn how they can be harnessed for a wide range of energy and environmental challenges.
“One of the biggest problems in the exploration of microbial dark matter is that so far, it is very complicated to determine when uncultured microbes are metabolically active and what their ecological function in a system is,” said study first author Roland Hatzenpichler, a postdoctoral researcher working in the Caltech lab of microbial ecologist Victoria Orphan. As part of the DOE Biological & Environmental Research (BER) program, Orphan’s lab has been studying the ecology and physiology of uncultured microorganisms catalyzing the anaerobic oxidation of methane in the environment. “When we deal with environmental or host-associated organisms that live within complex communities – typically thousands of species or more co-occur at the same time – it is very hard to determine what an individual species or cell is up to. Historically, this has been accomplished using single cell resolved isotope labeling. This is a comparatively tedious and expensive approach that is, however, very specific and sensitive. However, if we want to better understand microbiome functioning we need new, complementary approaches that are more high-throughput, that can be parallelized, and that are cheap. BONCAT [the new technique] is such a method.”
BONCAT: A High-Throughput Technique to Understand Microbiome Functions

This video shows the striking difference between the typical ocean seafloor and a methane seep site. As the submersible approaches the bottom of Hydrate Ridge, a seep site off the coast of Oregon, methane gas can be seen venting from below the seafloor. Atop the sediment, where a sample is being taken, a thick microbial mat has formed. Within this sediment, below the mat, clusters of microorganisms harvest energy by converting from methane by converting it into carbon dioxide. This video was taken during a research cruise of R/V Atlantis that was supported by the National Science Foundation (OCE-0825791). (Credit: Victoria J. Orphan, edited by Roland Hatzenpichler)
Short for BioOrthogonal Non-Canonical Amino acid Tagging, BONCAT is a technique developed at Caltech for bioengineering studies, but was adapted by Hatzenpichler and Orphan for use in microbial ecology investigations. BONCAT uses synthetic amino acids to label protein-making cells. These amino acids can then be stained with a fluorescent tag, lighting up active cells in their habitat. For this study, Hatzenpichler and his colleagues used sediment samples collected from deep-sea methane seep sediments off the coasts of Oregon and California for a series of incubation experiments, tracking the slow growth of methane-metabolizing bacterial and archaeal populations. The microbial communities in these sediments include aggregates of methane-oxidizing archaea called ANME (for ANaerobic MEthanotrophs) and sulfate-reducing bacteria (SRB) that live together symbiotically and help to remove some 80 percent of the methane released from ocean sediments.
The Caltech researchers combined BONCAT with fluorescent in situ hybridization (FISH) to analyze active microbes within these simulated environments and identify under which conditions they were active. “Typically, every methane seep site hosts several different groups of archaeal methane-oxidizers,” Hatzenpichler said. “A long-standing hypotheses has been that different members from these groups are differently active at different times. By exploiting the capability to do BONCAT analyses in comparatively high-throughput mode, we found members of all groups of methane-oxidizing archaea were anabolically active during the incubation. Now we can start asking why.”
Applying the BONCAT Technique to Flow Cytometry
DOE JGI researchers then helped develop a process that allowed the team to apply BONCAT to flow cytometry. “It’s the same process as for single cell genomics, but for aggregates of symbiotic bacteria and archaea,” said DOE JGI Microscale Applications Group head Rex Malmstrom of the technique called BONCAT-FACS (BONCAT – Fluorescence-Activated Cell Sorting). “We’re sorting individual aggregates full of cells that are active in a simulated environment and with the cell sorter we can grab active cells to study them. We had to figure out how to adapt the process for the flow cytometer. After the sorting, we performed whole genome amplification on individual aggregates then sequenced the 16S rRNA marker genes to identify cells comprising the aggregate. We’re now doing whole genome sequencing on these samples.” Sequencing could then provide researchers the genetic information explaining why some microbes were active under certain conditions, and why some others were not.

Caltech geobiologists who spearheaded the research: Victoria J. Orphan and Roland Hatzenpichler. (Credit: Stephanie Connon)
Through BONCAT-FACS, the team found that the methane-oxidizing archaea weren’t interacting with just the known sulfate-reducing bacteria, as had been expected, but were also sometimes associating with members of the less well-studied Verrucomicrobia phylum. These interactions had not been seen previously, and the finding suggests to researchers that methane-oxidizing archaea have a broader range of symbiotic relationships than had been thought. Whether these interactions are beneficial to both cell types or not is currently being addressed.
“JGI’s main power lies in the ability to decipher the genetic code of microorganisms,” noted Hatzenpichler. “By teaming up, we were able to isolate individual cell clusters involved in the degradation of the greenhouse gas methane from our samples, identify the cellular partners involved in this process, and gain insights into their genetic code.” Orphan and her team plan to use the genomic information from the paired methane-oxidizing archaeal and bacterial partners to develop deeper insights into the physiology and mechanisms of interaction and energetic exchange between different methanotrophic consortia coexisting in the environment.
In the fall, Hatzenpichler will be starting his own lab at Montana State University-Bozeman. He will continue to work closely with DOE JGI researchers to uncover the biogeochemical impact and biotechnological potential of uncultured microbes.
The work was supported by the NSF Center for Dark Energy Biosphere Investigations, the Gordon and Betty Moore Foundation, the Austrian Science Fund and the DOE’s Office of Science.