The Integrated Microbial Genomes group is focused on:
- Developing state-of-the-art data processing and analysis pipelines for the interpretation of microbiome omics data
- Developing forward-looking strategies for the deployment of computational workflows at peta- and exa- scales on multicore and manycore architectures with the ultimate objective of facilitating omics-based scientific investigations.
- Development and maintenance of the Integrated Microbial Genomes (IMG) data management system
Overall, the IMG group owns and maintains integral components of JGI’s production workflow, and is responsible for the annotation and analysis of (meta)genomic, (meta)transcriptomic, and functional genomic data and serving them to users via the Integrated Microbial Genomes (IMG) system (https://img.jgi.doe.gov).
Selected ongoing methods developed and related software resources are described below.
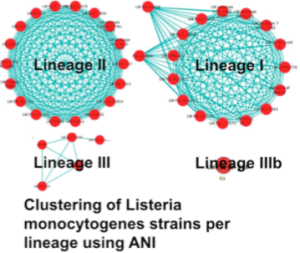 Average Nucleotide Identity (ANI)
Average Nucleotide Identity (ANI)
As part of this project, pairwise average nucleotide identities (ANI) and fraction of orthologous genomic regions (Alignment fraction, AF) have been computed for nearly 28,000 bacterial and archaeal genomes. By clustering genomes based on their pairwise AF and ANI values, we were able to ascertain mis-assignment of species names in genomes spanning nearly 18% of all existing species. Additionally, the complete linkage clustering made it possible to confidently assign species to nearly 326 genomes. Through the analysis of cliques, it has also become possible to identify speciation events within existing species. Within the JGI’s production pipeline, ANI is used to ascertain the species specificity of single cells and genomes extracted from metagenomes, and also as a metric for quality control within IMG. While ANI is integrated within IMG, http://ani.jgi-psf.org serves the current data.
Integrated Microbial Genomes (IMG)
The mission of the Integrated Microbial Genomes & Microbiomes (IMG/M) system is to support the annotation, analysis and distribution of microbial genome and microbiome datasets sequenced at DOE’s Joint Genome Institute (JGI). IMG/M is also open to scientists worldwide for the annotation, analysis, and distribution of their own genome and microbiome datasets, as long as they agree with the IMG/M data release policy and follow the metadata requirements for integrating data into IMG/M (see IMG/M submission site). The Integrated Microbial Genomes (IMG) system serves as a community resource for analysis and annotation of genome and metagenome datasets in a comprehensive comparative context. The IMG data warehouse integrates genome and metagenome datasets provided by IMG users with a comprehensive set of publicly available genome and metagenome datasets. IMG is a collaboration between the DOE JGI and the Biosciences Computing Group at the Computational Research Division of LBNL.
.
Research Team
 |
 |
 |
 |
|
|
I-Min (Amy) Chen
Lead, Computational Infrastructure
|
Neha Varghese, Software Developer | Marcel Huntemann, Software Developer |
Ken Chu Group Lead, Interface & Data Analysis |
|
| IMAChen@lbl.gov (925) 296-5697 |
njvarghese@lbl.gov (925) 296-5696 |
mhuntemann@lbl.gov (925) 927-2534 |
KLChu@lbl.gov (925) 926-5692 |
|
| Amy is leading the computational infrastructure groups of the Prokaryote Program. She is also leading the technical development of the Integrated Microbial Genomes (IMG) family of systems.
More about Amy here |
Neha’s research mainly focuses on the use of genome sequences to delineate prokaryotic organisms. She is also actively involved in expression analysis of transcriptomic and metatranscriptomic data, specifically exploring, benchmarking and implementing user-based RNAseq data analysis tools. | Marcel is in charge of several production pipelines (gene calling, functional annotation and methylomics) that run on microbial genomes and metagenomes. He also works on automating intra-department data exchange and assists with large scale computations on R&D projects. |
Ken is leading the development and maintenance of analytical tools and user interface components for the IMG family of systems. |
 |
 |
 |
 |
| Krishna Palaniappan,
Software Engineer, Database systems |
Manoj Pillay,
Software Engineer, Database systems
|
Jinghua (Jenny) Huang
Software Engineer, Database analysis |
Anna Ratner
Software Engineer, Database analysis |
| KPalaniappan@lbl.gov (925) 296-5710 |
mpillay@lbl.gov (925) 296-5820 |
JinghuaHuang@lbl.gov (925) 926-5827 |
ARatner@lbl.gov (925) 296-5823 |
| Krishna is responsible for the design and maintenance of the Integrated Microbial Genomes (IMG) data warehouse, and integration of genomic and metagenomic data with transcriptomics and proteomics data.
More about Krishna here. |
Manoj is responsible for integrating omics datasets into the IMG data warehouse, their distribution to users and the acquisition of public data from open-access sequence repositories into IMG.
More about Manoj here |
Jenny is working on the display and visualization of large amount isolate genomic and metagenomic data stored in Oracle databases, Berkeley DB and files in various formats.
More about Jenny here |
Anna is working on the display and visualization of large omics data. More about Anna here |
Selected Publications
- Ovchinnikov S. et al. (2017) Protein structure determination using metagenome sequence data. Science 355(6322):294-298
- Paez-Espino D. et al. (2017) IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 45(D1):D457-D465.Paez-Espino, D. et al. (2016) Uncovering Earth’s virome. Nature 536:425-30
- Chen IA. et al. (2017) IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45(D1):D507-D516.
- Paez-Espino, D. et al. (2016) Uncovering Earth’s virome. Nature 536:425-30
- Chen IM. et. al. (2016) Supporting community annotation and user collaboration in the integrated microbial genomes (IMG) system. BMC Genomics. 17:307
- Huntemann M. et al. (2016) The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4). Stand Genomic Sci. 11:17
- Huntemann M. et al. (2015) The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4). Stand Genomic Sci. 10:86.
- Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 2014 Jul 17;158(2):412-21. doi: 10.1016/j.cell.2014.06.034.
- IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Research 42 (Database-Issue): 568-573 (2014).
- IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Research 42 (Database-Issue): 560-567 (2014).