WALNUT CREEK, Calif.—A tiny crustacean that has been used for decades to develop and monitor environmental regulations is the first of its kind to have its genetic code sequenced and analyzed—revealing the most gene-packed animal characterized to date. The information deciphered could help researchers develop and conduct real-time monitoring systems of the effects of environmental remediation efforts.

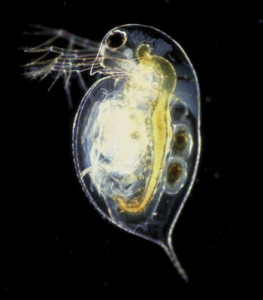
Daphnia pulex (commonly called water flea) Photo: Dr. Jan Michels, Christian-Albrechts-University, Kiel
Considered a keystone species in freshwater ecosystems, the water flea, Daphnia pulex, is roughly the size of the equal sign on a keyboard. Its 200 million-base genome was described in the February 4 issue of Science, the result of a collaboration between the Daphnia Genomics Consortium and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) that began nearly a decade ago when environmental agencies and toxicology researchers were looking into aquatic model systems that acted as sentinel species in order to diagnose the presence of problematic chemicals in fresh water and extrapolate their effects.
“Daphnia is one of the most widely used model systems for environmental protection agencies around the world,” said project leader and Indiana University Center for Genomics and Bioinformatics genomics director John Colbourne. “The costly challenge of evaluating conditions in the environment and of our water supplies may be overcome by Daphnia’s potential use as a high-tech and modern version of the mineshaft canary. An early detector of environmental risks should produce diagnostic and robust signals of the test-animal’s response to stress. Our initial studies revealed that Daphnia’s genes are evolved to be fine-tuned to environmental changes.”
The ability to use Daphnia for environmental studies focused on identifying associations between gene function and disease also contributed to the recent inclusion of Daphnia as a model system for biomedical research by the National Institutes of Health, joining established models like the fruit fly Drosophila and the worm C. elegans. Colbourne said that the Daphnia Genomics Consortium is working on improving techniques to do large-scale genome population assays to better track and manage remediation applications.
Aside from environmental monitoring, study coauthor Michael Pfrender of the University of Notre Dame noted another application of the Daphnia genome with regard to developing commercial biofuels. “When you grow algae in large open-air tanks to select for biofuel production, they’re invaded by Daphnia that graze down the algae,” he said. “You’re faced with either learning how to control Daphnia or learning how to use it to harvest the hydrocarbons.”
The Daphnia project has spawned a large research community that has grown over the course of the project from a handful of founding members to over 450 investigators distributed around the globe. Nearly 200 scientists have contributed published work resulting from the genome study.
The water flea was sequenced and annotated at the DOE JGI. Igor Grigoriev, head of the DOE JGI Eukaryotic Genomics group, noted that the Daphnia genome, which was sequenced using the Sanger method, is the most compact with over 31,000 genes, a third of which are of unknown function. Grigoriev attributed the large number of genes to expansion by tandem duplication, adding that the number is a conservative estimate. “There are more genes in Daphnia than there are in the human genome, more than any animal,” he said.
From the perspective of tracking a species’ evolution and development or phylogenetics, the Daphnia genome allows researchers a glimpse at a branch of the Arthropod phylum that had been left out until now when conducting comparative genomic studies without the group. While sequences of arthropods such as the honeybee (Apis mellifera) and the fruit fly (Drosophila melanogaster) are available, there was no representative from the crustaceans in the phylum.
“Crustaceans are the closest living relatives to insects, and Daphnia pulex is the first animal from this group to have its genome completely sequenced”, said Jeffrey Boore, who led the portion of this project done at the DOE Joint Genome Institute and is now CEO of Genome Project Solutions, which also made contributions to this study.
Annotating the Daphnia genome also afforded researchers an alternate way of determining gene functions. “This goes beyond having the first crustacean genome,” said Grigoriev. “By combining functional genomics assays we can see that the genes of unknown function – the new genes resulting from adaptations, not the ones conserved because they’re key for housekeeping – are the most responsive to environmental changes. In that sense, the organism’s environment can direct us to areas of interest; if you pick a genome and would like to annotate all the genes, go to the environment.”
The U.S. Department of Energy Joint Genome Institute, supported by the DOE Office of Science, is committed to advancing genomics in support of DOE missions related to clean energy generation and environmental characterization and cleanup. DOE JGI, headquartered in Walnut Creek, Calif., provides integrated high-throughput sequencing and computational analysis that enable systems-based scientific approaches to these challenges. Follow DOE JGI on Twitter.