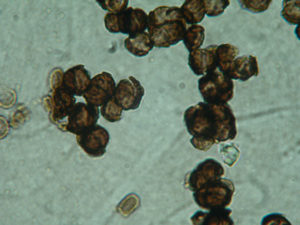
Black fungi are microscopic and mighty. They survive everywhere from Antarctica to Joshua Tree National Park, despite extremely harsh conditions. And their survival secrets could one day help other organisms survive hotter, drier climates. So University of Tuscia researchers Laura Selbmann and Claudia Coleine are working with scientists from around the world – and the JGI – to understand them better. Find show notes here.
Episode Transcript:
Menaka: Let’s begin this episode with a bit of a trip, to collect some microbe samples.
This journey takes us to a pretty remote place.We’re headed to the southern side of Antarctica, directly south of New Zealand, to an area called Victoria Land. There’s a mountain range that starts here, and runs through the continent. It’s called the transantarctic mountains. And that — is one place where these microbes live. So we’ve got to get over to those mountains, from a research base.
<<sounds of a helicopter>>
Menaka: <lightly yelling> And now, it gets a little loud! The only way to reach those mountains is by helicopter.
<<sounds of wind>>
Menaka: And once you land, it’s windy, for one thing. For another,
Laura Selbmann: It’s cold, full of snow.
Menaka: Laura Selbmann is the researcher who collects these kinds of samples. She works in a head to toe bright red snow suit, complete with goggles. Because it’s well below freezing. And the Antarctic landscape looks mostly empty and white. Snow covers nearly every surface in sight — but a few sandstone rocks peek out. And that’s where we’ll find what we’re after — inside those rocks. So it takes a few more tools to collect these samples.
<<sounds of a chisel banging into a rock>>
Menaka: A careful chisel opens up a whole little ecosystem, inside these rocks.
Laura Selbmann: Just below the rock surface, you see this blackish, this blackish band.

This rock is home to an entire microbial community: A black stripe of black fungi, and a green layer of phototrophs, as well as other viruses, archaea, and bacteria.
Menaka: It’s a little black squiggle running through the inside of the rock. You might mistake it for a different layer of rock, or even just a smudge of dark dirt. But that — is where the microbes are. So, Laura gathers the rock bits up in a plastic bag to bring back to the lab. Even in this unlikely place, these are microbes that are surviving.
<<music>>
Menaka: This is Genome Insider! Where we tag along with JGI-supported researchers who are studying all kinds of things and see how their work could advance clean energy and protect our environment.
I’m your host, Menaka Wilhelm.
And today, we’ve started off on the kind of sampling journey that Laura Selbmann makes. She’s based at the University of Tuscia, in Italy, studying botany and mycology, and specifically, these microbes that can exist in extreme environments. Like Antarctica, as Laura can tell you.
Laura Selbmann: The cold is something that you can expect, but, believe me, the, the main problem is the wind, because if you don’t have wind, even if you are at minus 30, for instance,
Menaka: That’s Celsius, about minus 20 Fahrenheit –
Laura Selbmann: You can work because the climate is very dry.
Menaka: That still sounds very cold, even without wind! But — that dry climate is another key to what makes these Antarctic microbes so interesting. It’s not just that they weather extremely cold temperatures. They also live with very little water. Because below freezing, there’s not much water around. It’s essentially all locked away in ice and snow.
And still, these microbes persist. And not only do they persist. They operate as a little microbial community, even with very few of the basic ingredients for an ecosystem. Within that community, you find all kinds of microbes — algae, cyanobacteria, bacteria, viruses.
But there’s one kind of microbe — a fungus — that stands out. It’s what Laura pointed out when she found that black squiggle inside the Antarctic rock. That black color is also where these fungi get their name.
Laura Selbmann: Black fungi are, really, the excellence in the extremes.
Menaka: These fungi stand out for their ability to break things down, and make new chemicals. Similar to how forest fungi, like mushrooms, grow on decomposing trees, and return nutrients to the forest floor, these microscopic black fungi can do similar recycling — just at a teeny tiny scale.
Laura Selbmann: So they guarantee carbon, nitrogen cycles, even in these remote, uh, environments where few organisms can survive.
Menaka: And understanding how these fungi pull that off could eventually be useful for other organisms around the world, especially as climate change makes more places hotter and drier. So to understand how these fungi can bravely cycle on, despite a very barren environment, Laura is taking a look at their genomes. She’s looking beyond just Antarctica — because, as you might imagine, these hardy black fungi also live in plenty of other harsh places all around the world.
<music>
Menaka: From Joshua Tree National Park in Southern California, to Nepal’s Himalayan mountains. In contaminated places, like the super acidic, heavy metal laden Rio Tinto in Spain, and at nuclear waste sites like the remains of the Chernobyl nuclear power plant. Even in the cracks of marble monuments, where these fungi survive on the nutrients that are provided in air pollution.
For these little black fungi, what a track record! They’re ready to face high levels of salt, super low water, extreme temperatures, both high and low, chemical and nuclear contamination, and access to almost no nutrients.
So Laura wants to know which genes let them survive all of that. She’s partnered with the JGI to sequence and analyze these fungal genomes. And she’s also working with 19 other research groups from around the world. They’ve picked fungi from all kinds of places, including many of the harshest environments that nature has to offer. And they’ll sequence lots of samples to build reference genomes. They’ll do that for 92 forms of black fungi in total —along with smaller studies of hundreds of other species.
And there’s a reason the JGI is interested in so many different kinds of black fungi. Despite their shared name, and hardiness, these organisms aren’t all the same. Far from it!
Laura Selbmann: These organisms are phylogenetically totally unrelated because we find black fungi, general definition, in the whole fungal tree of life.
Menaka: So these fungi are all different. They’ve all evolved along different paths.
Laura Selbmann: But if you look at them at the microscope, they look like, almost the same.
Menaka: They grow in similar patterns,
Laura Selbmann: Sort of cauliflower colonies, so growth in all directions.
Menaka: But mainly, they’ve got coloring in common.
Laura Selbmann: All of them are black.
Menaka: And that’s probably key for their hardiness. Because even after evolving separately, all of these black fungi get their color from the same molecule — melanin. It’s the same kind of pigment that colors our skin, but not exactly the same molecule. And Laura is particularly interested in how these fungi make and maintain this melanin — the molecule that brings them all together.
Melanin may even be interesting in ways we don’t understand yet. But here’s one place we know it’s working to black fungi’s advantage. In nuclear waste sites, black fungi, and anything else around gets hit with something called ionizing radiation. Which, for most living things, is a real threat. But for black fungi, melanin provides a shield. In fact, because of that melanin, black fungi can even absorb the radiation’s energy, process it, and use that energy themselves.
Laura Selbmann: What is normally dangerous, ionizing radiation, and what is normally, harmful to life, can be converted, but to this organism in something that allows them to enhance growth.
Menaka: So one day, black fungi might actually help us break down this kind of waste. Because not only can these microbes withstand this radiation, they use it to thrive, thanks to that melanin.
And that’s just one way that they’re world-class survivors. From their DNA, their genomes, we have a lot more to learn about how they live their very extreme lives.
Laura Selbmann: For now, we don’t have the awareness of exactly what are the abilities, the genomic abilities of these organisms. What is into the genomes that make them so special in the extremes?
Menaka: And of course, to answer that question, you’ve got to take a look at a lot of fungal DNA! Which is where the JGI comes in – first, with sequencing, then assembling and annotating those genomes. Laura and her team will also be testing RNA and compounds these fungi make in transcriptomics and metabolomics experiments, to learn even more about these fungi. But there are a few challenges to doing that work — because many of the same traits that make black fungi super rugged, also make these microbes tough to handle in test tubes. We’ll get into that after the break.
<<music>>
Menaka: First, it’s time to guess a JGI acronym. Because we’ve got a lot of them. And the question is, how guessable are they? Today, I’ll be quizzing Amber Golini.
Menaka: So, before we get to our acronym – what do you do at the JGI, Amber?
Amber Golini: I’m a research associate, in Trent Northen’s metabolomics lab. I work on sample processing and preparation for metabolomics research.
Menaka: Cool. And what does that entail?
Amber Golini: So my main tasks are to prepare, extract, and process samples to be run on our instrumentation systems. So this involves setting up and operating the instruments, trouble-shooting and calibrating the systems, as well as monitoring the sample runs to make sure the data is collected properly.
Menaka: Excellent. Well, we’ve got a great acronym for you to guess today. It’s IMG. So IMG. And here are your options. Does IMG stand for,
- Integrated Microbial Genomes
- International Miscanthus Group
- Investigating Mutant Grass
- Identification of Microalgae Genetics
Menaka: Do you think it’s 1, 2, 3 or 4?
Amber: I’m going to go with 1 on this one.
Menaka: Nice. Let’s hear from our revealer.
Rekha Seshadri: Hi Menaka, I’m Rekha, and I’m a computational biologist at the Joint Genome Institute. So IMG stands for Integrated Microbial genomes!
Menaka: Great work, you guessed correctly! So Integrated Microbial Genomes is a data portal that anyone can access online. Had you heard of it before?
Amber Golini: I actually had not!
Menaka: Wow. Well, an even better guess then. Here’s a bit more about that project from Rekha Seshadri.
Rekha: So we have a whole collection of bacterial and archaeal isolate genomes, we have environmental samples, metagenomic sequences, single cells, and, so you can gain access, you can do smart searches against all of that data. And then once you find the data that you’re interested in, you can ask biological questions and answer them.
Menaka: And you can find that integrated microbial genomes database on our website. At img.jgi.doe.gov. We’ll link to it in the show notes, too. But that’s img.jgi.doe.gov.
Before we get back to the black fungi — a bit more about the nuts and bolts of this project.
<<music>>
Menaka: This work on black fungi is specifically called Shed LighT in the daRk lineagES of the Fungal tree of life — shortened to STRES. And the JGI supports it via the Community Science Program, where we accept new projects every year.
You can find out more about that program, and how to submit your own proposal, at our website. There’s a link in the show notes.
<<music>>
Menaka: OK — Now, back to the black fungi. As a quick recap — these fungi are interesting because they’re resilient. In the face of dry conditions, and super hot or cold temperatures, even contamination. If you can think of a harsh environment, there’s probably black fungi happily going about their life there.
And these fungi are black because of melanin pigments. These pigments seem to be part of the recipe of what’s making these fungi so tough, but there’s lots more to find out. So Laura Selbmann, and her collaborators, are heading to different sites around the world, collecting bits of rock and other samples with black fungi, and bringing them back to the lab.
This is where things get a bit tricky — because a black fungus is prepared to survive many difficulties, as we’ve established. However, cooperating in the tame, fluorescent environment of microbiology research requires a totally different skillset. Black fungi have all kinds of traits that help them in life, not in lab.
But Laura and her team need these fungi to cooperate to get at their genomes. These teams are careful to collect small samples from delicate environments, but genetic sequencing only works accurately if you have enough DNA for machines to read. So before sequencing, they have to get these wild black fungi to grow.
Claudia Coleine is a postdoc who works with Laura. She told me how this starts, from a powdered rock sample.
Claudia Coleine: The first thing we did we do is, spread the powdered rocks on a plate, composed of multiset agar.
Menaka: That agar provides nutrients for the fungus to grow — which it does, on its own terms. Here’s Laura, again.
Laura Selbmann: One of the main challenges to working with these organisms is that they are very, very slow growing, incredibly slow growing.
Menaka: Growth requires nutrients and water — both of which are in short supply in this fungus’s usual life. So growing extremely slowly is a useful adaptation for extreme conditions, where the supply budget is tight. But this also means it’s very hard to coax these fungi onto a research timeline.
Claudia Coleine: Some Antarctica species take, around three or four months to get a good quantity of biomass, for DNA extraction.
Menaka: And once there is enough fungi grown, there’s another obstacle standing between these researchers and these fungal genomes, because again, they’ve evolved for extremes, not experiments. Black fungi have a cell wall protecting each of their cells that’s quite thick, and tough, because of the melanin inside. So it’s difficult to break open those cells, to separate out their DNA.
Laura Selbmann: And this of course, affects the procedures to get a good amount of DNA and DNA of good quality.
Menaka: So Laura and Claudia, along with their other collaborators, have been testing lots of ways to separate out that DNA, and working with the JGI to be sure there are samples that work for all the sequencing processes.
Claudia Coleine: Moreover JGI will help us to perform, if needed, species-specific DNA or RNA extraction protocols.
Menaka: Because there are lots of species to sequence! As this project goes on, they’re taking a look at black fungi from all over the planet.
Claudia Coleine: One aim of the project is to compare Antarctica with the hot desert and see if the microbial community composition is shared or is very different according to the different conditions, according to the geography and different environmental conditions.
Menaka: Claudia’s sense is that some fungal species may be able to exist across many different desert conditions — kind of a core group of desert-weathering fungi.
Claudia Coleine: But I also expect to find different community composition across deserts. Especially according to the water conditions.
Menaka: And maybe most importantly, this project will survey black fungi broadly enough to get a real sense of lots of different species.
Claudia Coleine: With this project JGI will help us to increase, to expand, the catalog of black fungi genomes, of more than 600 genomes.
Menaka: So this project will give us a look at the survival traits from hundreds of species in many different environments. And across all of these fungi, more genomic information will help us understand where those traits come from. With all of their experience in harsh environments, black fungi might have a lot to teach all of the life forms living in cushier places on our planet.
<<music>>
Menaka: That wraps up this journey to the center of these Antarctic rocks! We’ll be back in a month. We’ll cover something closer to the JGI itself – a tour of how samples go from shipments to genetic sequences.
This episode was written and produced by me – Menaka Wilhelm, with production help from Massie Ballon, Allison Joy, and Ashleigh Papp.
Many thanks to Laura Selbmann, Claudia Coleine, and the Italian National Program for Antarctic Research, funded by the Italian Ministry of University and Research.
If you liked this episode, help someone else find it! Tell them about it, send a link over, or leave us a review wherever you’re listening to the show.
Genome Insider is a production of the Joint Genome Institute, a user facility of the US Department of Energy Office of Science located at Lawrence Berkeley National Lab in Berkeley, California.
Thanks for tuning in – until next time!
Show Notes
- Episode Transcript
- How Black Fungi Adapt to Extremes
- Integrated Microbial Genomes and Microbiomes
- Submit a proposal to work with the JGI
- Our contact info:
- Twitter: @JGI
- Email: jgi-comms at lbl dot gov
Genome Insider is a production of the Joint Genome Institute.
Audio in the opening scene comes from an actual expedition Laura took to Antarctica. Laura.Selbmann©PNRA
All the sampling activities in Antarctica have been performed in the frame of italian expeditions of the Italian National Program for Antarctic Researches (PNRA), funded by the Italian Ministry of University and Research; all specimens collected and fungi isolated are preserved in the Culture Collection of Fungi from Extreme Environment, the Italian National Antarctic Museum (MNA-CCFEE)