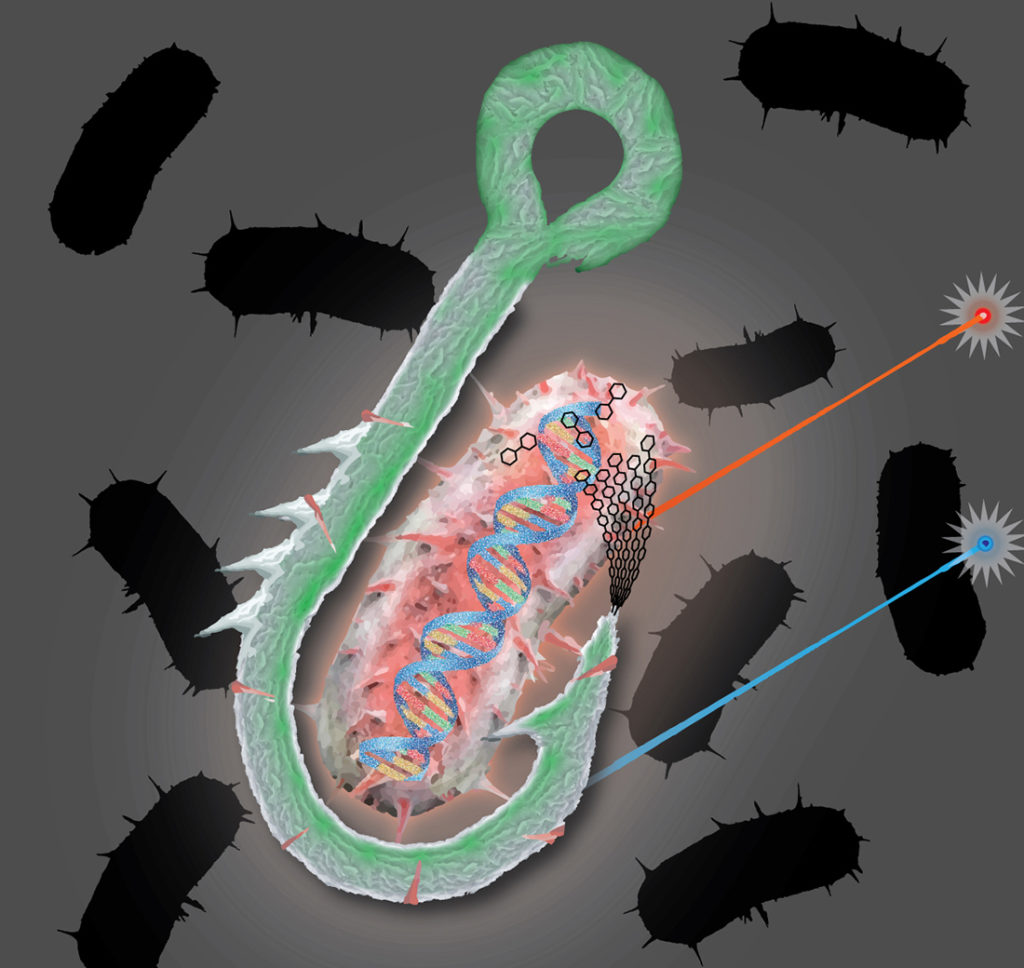
A “bait and hook” single-cell genomic approach to bioprospecting.
The Science
One of the most vital pieces of equipment for fly fishing is a boxful of lures. Designed with feathers or wires to mimic an insect or a particular movement, each of these lures are the bait designed to attract specific catches. A similar technique has been developed by researchers led by Tanja Woyke at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab). Using a single-cell screen, they can now identify microbes with specific functional characteristics. When they tested the screen method on a microbial community from geothermal hot springs, they uncovered a novel cellulose-degrading bacterium typically found in low abundances.
The Impact
Microbes are known to be key contributors to the planet’s biogeochemical cycles, but the vast majority remains uncultivated and unknown. The challenge remains to learn more about what microbes are present in any given ecosystem—particularly those that are in low abundances—and their functional roles in their community. This study shows how the function-driven, single-cell screen can uncover more about cellulose-degrading microbes. The new technique is another tool for microbial ecologists to learn more about the diversity of microbial enzymes, which could have a plethora of potential applications to industrial processes.
Summary
JGI researchers developed a function-driven, single-cell screen that they described as a “bait and hook” approach to isolate cellulose-degrading microbes. They first bound single microbes to fluorescently labeled cellulose particles that served as the “bait,” and then they isolated the particles through the “hook” of fluorescence-activated cell sorting to sort cells into several small pools, followed by whole genome amplification and shotgun sequencing. The team first tested the approach on bacterial laboratory models to set a reference for its performance, and then applied it to environmental samples from the Great Boiling Spring Geothermal Field in Nevada. To validate the predicted cellulases from their captured genomes, the team then used DNA synthesis and cloning, followed by protein in vitro expression and mass spectrometry-based enzyme activity assay. The work was reported in The ISME Journal.
While the screen caught several microbes from known cellulose-degrading groups, the researchers also hooked and were able to reconstruct a high-quality draft co-assembled genome for a new genus level member of the novel candidate phylum Goldbacteria. They’ve proposed the name Candidatus ‘Cellulosimonas argentiregionis’ for this microbe.
The work is an example of how function-driven genomics, which Woyke and her colleagues define as “any form of functional enrichment leading to the sequencing of genomes from either individual cells or consortia of cells,” can be applied. The approach can be further developed to bait for specific functions other than cellulose-degradation to tease out uncultivated microorganisms of interest from complex environmental samples.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Tanja Woyke
Microbial Program Head
DOE Joint Genome Institute
[email protected]
Funding:
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231. The work was supported by a JGI Director’s Science award. The DOE Joint BioEnergy Institute (http://www.jbei.org) is supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. Support for this research was also provided by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under contract DE-FC02-07ER64494. EG is supported by the NIGMS Biotechnology Training Program (NIH 5 T32 GM008349) at the University of Wisconsin – Madison. This work was also supported under the LBNL Microbes to Biomes Laboratory Directed Research and Development program (LDRD) entitled ‘Tackling microbial-mediated plant carbon decomposition using “function-driven” genomics’.
Publication:
- Doud DFR et al. Function-driven single-cell genomics uncovers cellulose-degrading bacteria from the rare biosphere. ISME J. 2019 Nov 21. doi: 10.1038/s41396-019-0557-y.
By Massie S. Ballon