A collaborative team led by researchers at the University of Texas (UT) at Austin, the HudsonAlpha Institute for Biotechnology (HudsonAlpha), and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility, has discovered that the candidate bioenergy feedstock switchgrass has adapted to expand its habitat range, but at what cost? In the following guest piece, postdoctoral researcher Joseph Napier from the Juenger lab and computational biologist Paul Grabowski of HudsonAlpha outline the questions they asked – and answered – in the study that recently appeared in the Proceedings of the National Academy of Sciences (PNAS).
 Switchgrass, Panicum virgatum, is a widespread, perennial grass in North America that has been the target of extensive natural sampling and field experiments due to its potential for habitat restoration and as a feedstock for biomass-based biofuel production. As seen with many commonly cultivated commercial crops such as wheat, peanut, oat, banana, potato, etc., switchgrass is polyploid. This means it contains more than two copies of the genome, which can have pronounced ecological and evolutionary consequences.
Switchgrass, Panicum virgatum, is a widespread, perennial grass in North America that has been the target of extensive natural sampling and field experiments due to its potential for habitat restoration and as a feedstock for biomass-based biofuel production. As seen with many commonly cultivated commercial crops such as wheat, peanut, oat, banana, potato, etc., switchgrass is polyploid. This means it contains more than two copies of the genome, which can have pronounced ecological and evolutionary consequences.
A previous study conducted by our research team surveying the genomic and phenotypic diversity across the natural range of switchgrass found that polyploidy has enhanced the adaptive potential of switchgrass and is a key component in providing genetic variation available for selection to improve biofuel yield through a changing future. Switchgrass has multiple naturally occurring ploidy levels, called cytotypes, across the species. These are primarily tetraploids (4X or four copies of the genome) and octoploids (8X or eight copies of the genome).
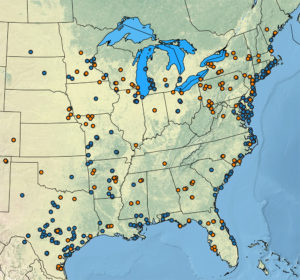
Documented occurrences of different switchgrass cytotypes (4X in blue and 8X in orange) throughout the United States. One of the early interests in exploring 8X switchgrass was because the noticeable occurrence of 8X in 4X distribution gaps. (Joseph Napier)
Lovell et al. focused on the 4X variety, largely because it’s been prioritized in breeding efforts, and also because 8X switchgrass is generally thought to harbor less genetic and phenotypic (visible traits) diversity. After our team finished analyzing the diversity present in 4X switchgrass, we were surprised by the number of 8X individuals that had been collected from field work. When viewing the full distribution of switchgrass on a map, these 8X individuals occurred in parts of the switchgrass range where 4X were noticeably absent (see figure on the right).
This pattern challenged the idea that 8X switchgrass was an unremarkable, homogenous group and raised several important questions. Where did the 8X variant come from? Did they arise multiple times or does their broad distribution indicate multiple, independent origin events? Do the cytotypes react differently in response to the environment? We set out to explore these questions and solve the mystery behind the observed 8X switchgrass patterns. In the process, we also hoped to use this unique opportunity to quantify how transitions to higher ploidy could be generating these observed patterns. As many other plant species have natural cytotype variation, we also hoped that this would more broadly allow us to assess how ploidy variation alters genomic diversity, fitness, and adaptability.
Using a combination of genomic, quantitative genetic, landscape, and niche modeling approaches, we contrasted the diversity of 4X and 8X switchgrass across hundreds of naturally-occurring genotypes (in this case, a plant’s complete genetic makeup) and 10 common gardens. We discovered that 8X populations have arisen multiple times from different genetic backgrounds, and that these 8X populations contain novel combinations of genetic diversity. We also found that much of the variation in physical characteristics that is seen in 4X switchgrass is also observed across the 8X cytotype. However, the 4X and 8X cytotypes diverge in their response to climate variations between the common gardens, indicating a generalist (8X)-specialist (4X) tradeoff. Furthermore, niche modeling suggests niche evolution between 4X and 8X linked to climate adaptation. Overall, these results indicate that the 8X represent a unique combination of genetic variation that has allowed the expansion of switchgrass’ ecological niche. The knowledge gained from 8X switchgrass is a valuable resource towards the effort to generate climate resilient switchgrass for bioenergy production.
Building on the framework provided by our new switchgrass manuscript, we plan to use existing data as well as generating additional genomic resources to address long-standing evolutionary ecological hypotheses about how C4 perennial grasses like switchgrass respond to pronounced climate shifts. (C4 plants use a more efficient form of photosynthesis that reduces water loss, compared to the process used by most plant species.) Understanding how the interplay between gene flow, population statistics, and adaptation mediated the response of switchgrass to past climate change will provide unprecedented insight into the mechanisms that perennial grasses have and will use to respond to climate shifts. Changes in ploidy might play a key role in this response.
Generating additional genomic resources for the 8X will provide a rare opportunity to assess the generality of a “ploidy hop,” when switchgrass (and by extension similar taxa) are faced with pronounced environmental fluctuations. New findings from this line of research combined with past insights will allow the identification of potentially novel combinations of genetic diversity present in switchgrass that might be linked to climate adaptation and range expansion. This would subsequently represent a valuable breeding resource for expanding the range of suitable growing conditions and enhancing the resilience of switchgrass feedstock production.
Publication:
Napier, J. D., Grabowski P.P., et al. (2022) A generalist-specialist tradeoff between switchgrass cytotypes impacts climate adaptation and geographic range. Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.2118879119
References:
- HudsonAlpha story for PNAS paper: More genome copies in switchgrass leads to increased climate flexibility
- Lovell, JT et al. (2021). Polyploidy and genomic introgressions facilitate climate adaptation and biomass yield in switchgrass. Nature, 590(7846), 438-444.
- JGI Release: Fields of Breeders’ Dreams: A Team Effort Toward Targeted Crop Improvements
- JGI Video: The Importance of Having a Switchgrass Genome
- JGI Video: Origins of the Switchgrass Common Gardens
- JGI Podcast: Exploring the Diversity of the American Prairie’s Switchgrass