My project at the Joint Genome Institute (JGI) was to measure the polymorphisms that transposable elements (TEs) contributed to in the grass model species Brachypodium hybridum. This is a relative of the JGI Flagship Plant B. distachyon. TEs are DNA sequences that move along the genome and they can create genetic variation in a population. Genetic variants that involve the gain or loss of a DNA sequence are called presence/absence polymorphisms.
This work is a study on TEs to understand how they contribute to the evolution of a polyploid species, which include more than two sets of chromosomes. Many crop species are polyploids, like wheat and oats, which allows our study to be used as a model for these crops as well as for grass species that are polyploids. TE research in polyploids will allow researchers to alter the DNA of plant species to improve them. For example, the research could be used to aid plant breeding. The information gathered by this study will help to understand the basic biology of the polyploid grasses (for example, switchgrass, wheat, sugarcane, etc.), and eventually lead to the improvement of biofuel grasses or food grasses.
This marked my second year interning with the JGI. After my first summer internship (in 2020), I was offered a position as a student assistant in John Vogel’s Plant Functional Genomics research group to continue working on the project. I then decided to extend this work as an intern again for the second summer. The difficulty of my work varied drastically throughout these positions due to different tasks requiring different levels of skill and knowledge, as well as the number of tasks increasing over time. However,I was able to adapt and learn with the help of my mentors.
Before beginning my internship with JGI for my first summer in 2020, my coding experience was limited to R-studio and Python, whereas this project worked mostly in Linux and focused on plant genome research. My mentors, Dr. Li Lei and Ms. Virginia Scarlett, were able to answer most of my questions and pointed me in the direction of solutions if they did not have the answers themselves.
Due to the research fully being online, I faced many challenges, from hardware issues to feeling disconnected from the research itself. Although the challenges were difficult, I adapted quickly to online research by asking my mentors many questions and by looking for real world applications of my project.
Charting Growth over Time
At the end of my first summer internship, I was able to obtain results by coding with shell scripting and I plotted the data with R.
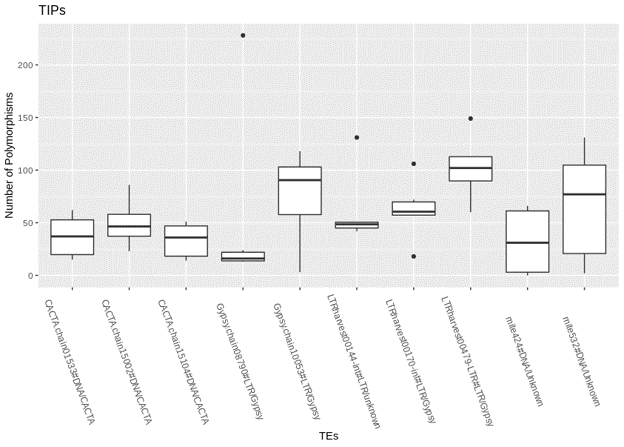
The graph shows that multiple TEs contribute to the majority of TE Insertion Polymorphisms (TIPs) in Brachypodium hybridum accessions and that the number of TIPs contributed vary between the TEs.
As a student assistant, I worked to further understand the effect of TEs on Brachypodium hybridum, created code on R-studio for plotting data gathered from shell scripts, and I organized accession information on the Brachypodium genus on an excel sheet.
During my second summer internship at JGI, I massively improved my skills and was able to obtain much better results and display them more clearly.
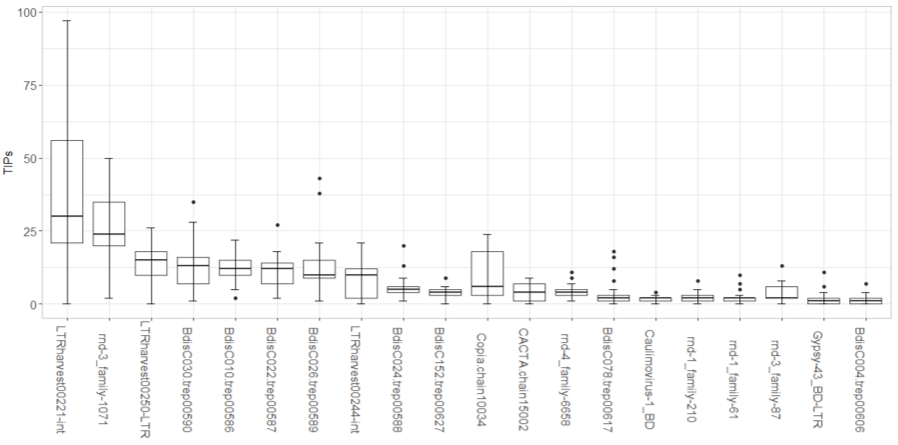
The new boxplots show that transposable elements (TEs) vary in their contribution to TE insertion polymorphisms (TIPs) and that several TEs contribute to a majority of the TIPs in Brachypodium hybridum accessions. We also see an overlap in the eight TEs that contribute the most to Brachypodium hybridum and its parent species, Brachypodium distachyon (BdiscC030, BdisC010, BdisC022, BdisC026).
After creating the new boxplot this summer, my mentors and I wanted to figure out the percentage of TIP contributions from TEs. So we plotted a bar graph that shows the percentage of TIPs that each TE contributes to.

In this graph, the percentages are very similar across the different accessions of Brachypodium hybridum, which was also seen in Brachypodium distachyon.
One of the largest challenges I faced during my internships at the JGI was figuring out how to communicate my issues properly so that my mentors could help. In addition to learning new tools and coding techniques, I also had to develop a new vocabulary in bioinformatics. All of this required much time and help from my mentors to ensure I learned them correctly. Without this opportunity and my mentors, I never would have been given the skills and confidence to follow through with my decision to apply to graduate school in the bioinformatics field.
Written by: Jacob Espinosa
Espinosa is a Bioengineering Senior at the University of California, Merced.
